Farhan Tanvir
Tackling Oversmoothing in GNN via Graph Sparsification: A Truss-based Approach
Jul 16, 2024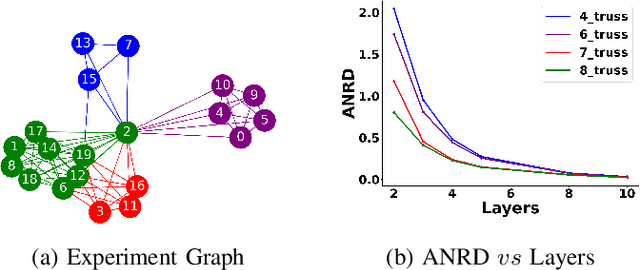
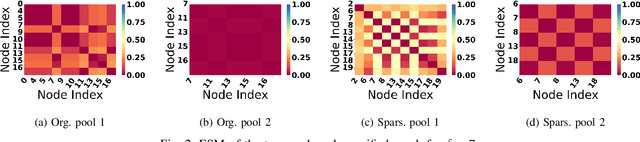
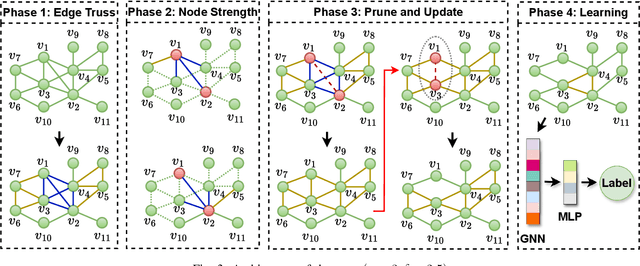
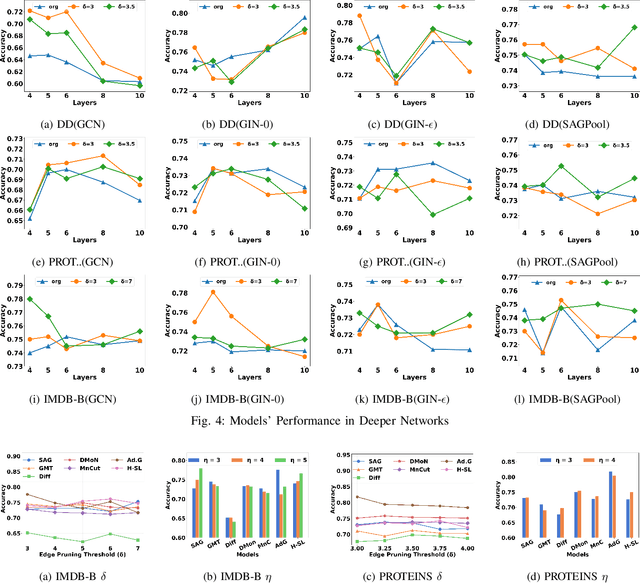
Abstract:Graph Neural Network (GNN) achieves great success for node-level and graph-level tasks via encoding meaningful topological structures of networks in various domains, ranging from social to biological networks. However, repeated aggregation operations lead to excessive mixing of node representations, particularly in dense regions with multiple GNN layers, resulting in nearly indistinguishable embeddings. This phenomenon leads to the oversmoothing problem that hampers downstream graph analytics tasks. To overcome this issue, we propose a novel and flexible truss-based graph sparsification model that prunes edges from dense regions of the graph. Pruning redundant edges in dense regions helps to prevent the aggregation of excessive neighborhood information during hierarchical message passing and pooling in GNN models. We then utilize our sparsification model in the state-of-the-art baseline GNNs and pooling models, such as GIN, SAGPool, GMT, DiffPool, MinCutPool, HGP-SL, DMonPool, and AdamGNN. Extensive experiments on different real-world datasets show that our model significantly improves the performance of the baseline GNN models in the graph classification task.
HeTriNet: Heterogeneous Graph Triplet Attention Network for Drug-Target-Disease Interaction
Nov 30, 2023
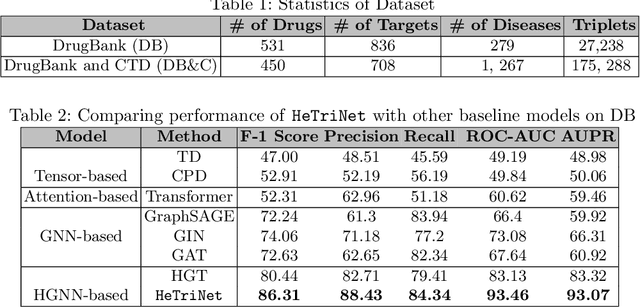
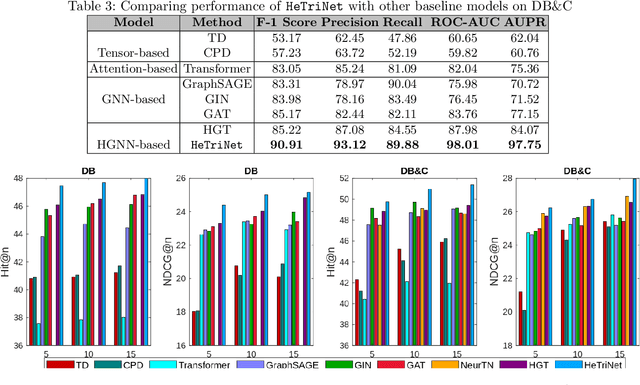

Abstract:Modeling the interactions between drugs, targets, and diseases is paramount in drug discovery and has significant implications for precision medicine and personalized treatments. Current approaches frequently consider drug-target or drug-disease interactions individually, ignoring the interdependencies among all three entities. Within human metabolic systems, drugs interact with protein targets in cells, influencing target activities and subsequently impacting biological pathways to promote healthy functions and treat diseases. Moving beyond binary relationships and exploring tighter triple relationships is essential to understanding drugs' mechanism of action (MoAs). Moreover, identifying the heterogeneity of drugs, targets, and diseases, along with their distinct characteristics, is critical to model these complex interactions appropriately. To address these challenges, we effectively model the interconnectedness of all entities in a heterogeneous graph and develop a novel Heterogeneous Graph Triplet Attention Network (\texttt{HeTriNet}). \texttt{HeTriNet} introduces a novel triplet attention mechanism within this heterogeneous graph structure. Beyond pairwise attention as the importance of an entity for the other one, we define triplet attention to model the importance of pairs for entities in the drug-target-disease triplet prediction problem. Experimental results on real-world datasets show that \texttt{HeTriNet} outperforms several baselines, demonstrating its remarkable proficiency in uncovering novel drug-target-disease relationships.
Seq-HyGAN: Sequence Classification via Hypergraph Attention Network
Mar 19, 2023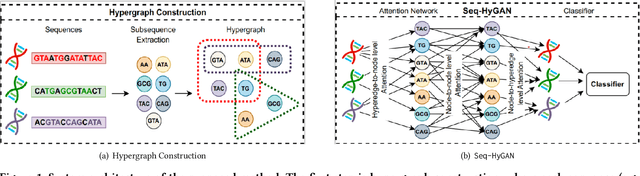
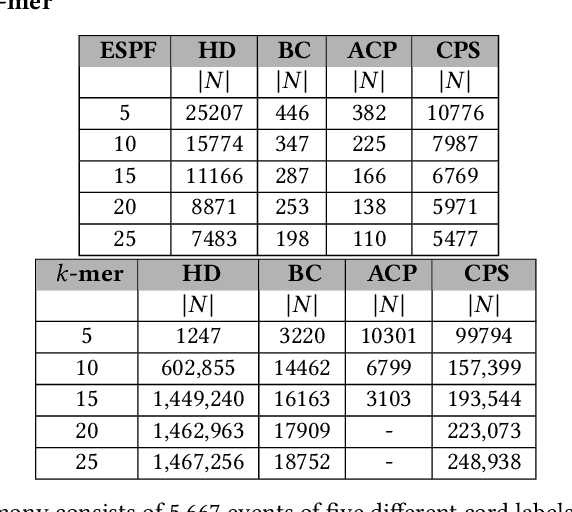
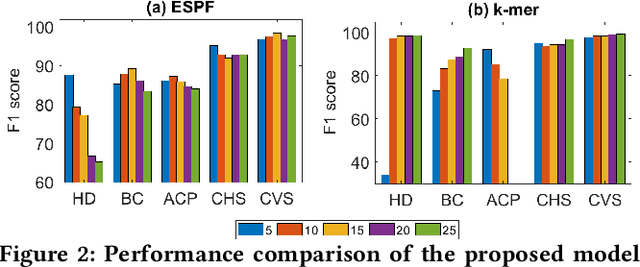
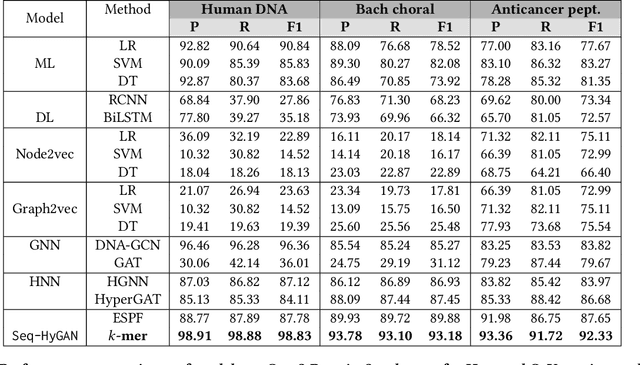
Abstract:Sequence classification has a wide range of real-world applications in different domains, such as genome classification in health and anomaly detection in business. However, the lack of explicit features in sequence data makes it difficult for machine learning models. While Neural Network (NN) models address this with learning features automatically, they are limited to capturing adjacent structural connections and ignore global, higher-order information between the sequences. To address these challenges in the sequence classification problems, we propose a novel Hypergraph Attention Network model, namely Seq-HyGAN. To capture the complex structural similarity between sequence data, we first create a hypergraph where the sequences are depicted as hyperedges and subsequences extracted from sequences are depicted as nodes. Additionally, we introduce an attention-based Hypergraph Neural Network model that utilizes a two-level attention mechanism. This model generates a sequence representation as a hyperedge while simultaneously learning the crucial subsequences for each sequence. We conduct extensive experiments on four data sets to assess and compare our model with several state-of-the-art methods. Experimental results demonstrate that our proposed Seq-HyGAN model can effectively classify sequence data and significantly outperform the baselines. We also conduct case studies to investigate the contribution of each module in Seq-HyGAN.
DDI Prediction via Heterogeneous Graph Attention Networks
Jul 12, 2022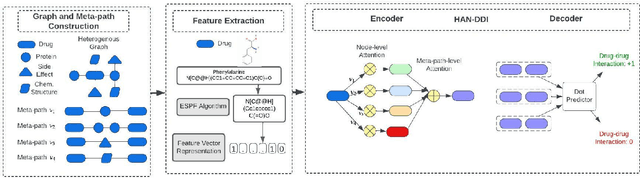
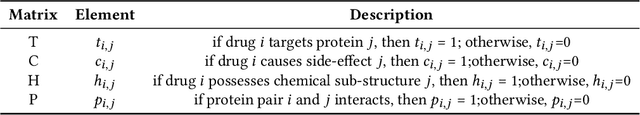

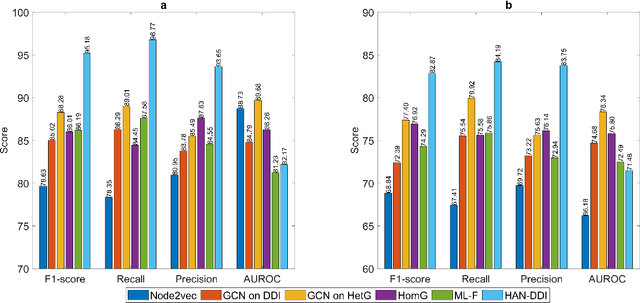
Abstract:Polypharmacy, defined as the use of multiple drugs together, is a standard treatment method, especially for severe and chronic diseases. However, using multiple drugs together may cause interactions between drugs. Drug-drug interaction (DDI) is the activity that occurs when the impact of one drug changes when combined with another. DDIs may obstruct, increase, or decrease the intended effect of either drug or, in the worst-case scenario, create adverse side effects. While it is critical to detect DDIs on time, it is timeconsuming and expensive to identify them in clinical trials due to their short duration and many possible drug pairs to be considered for testing. As a result, computational methods are needed for predicting DDIs. In this paper, we present a novel heterogeneous graph attention model, HAN-DDI to predict drug-drug interactions. We create a heterogeneous network of drugs with different biological entities. Then, we develop a heterogeneous graph attention network to learn DDIs using relations of drugs with other entities. It consists of an attention-based heterogeneous graph node encoder for obtaining drug node representations and a decoder for predicting drug-drug interactions. Further, we utilize comprehensive experiments to evaluate of our model and to compare it with state-of-the-art models. Experimental results show that our proposed method, HAN-DDI, outperforms the baselines significantly and accurately predicts DDIs, even for new drugs.
HyGNN: Drug-Drug Interaction Prediction via Hypergraph Neural Network
Jun 25, 2022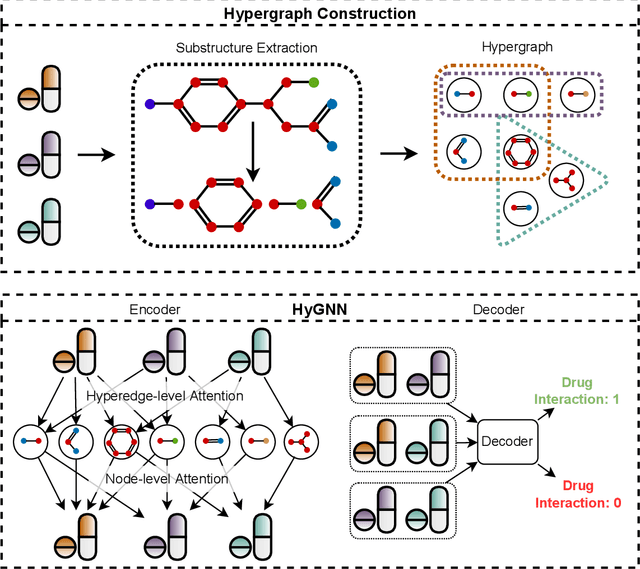
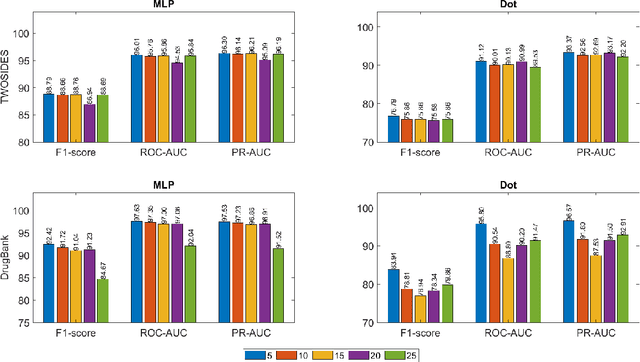
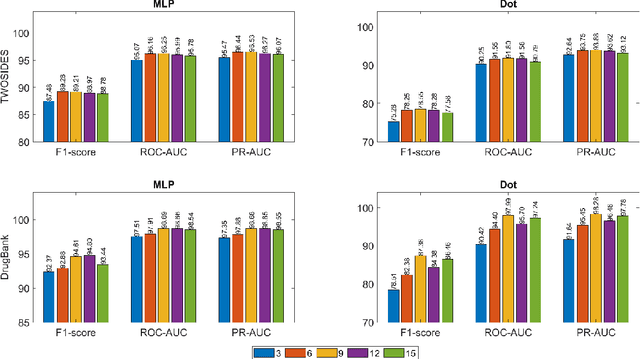
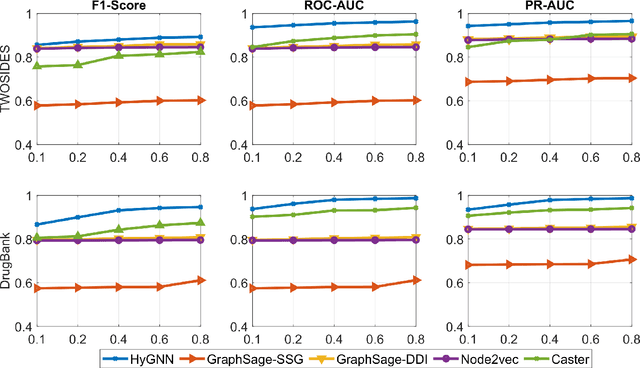
Abstract:Drug-Drug Interactions (DDIs) may hamper the functionalities of drugs, and in the worst scenario, they may lead to adverse drug reactions (ADRs). Predicting all DDIs is a challenging and critical problem. Most existing computational models integrate drug-centric information from different sources and leverage them as features in machine learning classifiers to predict DDIs. However, these models have a high chance of failure, especially for the new drugs when all the information is not available. This paper proposes a novel Hypergraph Neural Network (HyGNN) model based on only the SMILES string of drugs, available for any drug, for the DDI prediction problem. To capture the drug similarities, we create a hypergraph from drugs' chemical substructures extracted from the SMILES strings. Then, we develop HyGNN consisting of a novel attention-based hypergraph edge encoder to get the representation of drugs as hyperedges and a decoder to predict the interactions between drug pairs. Furthermore, we conduct extensive experiments to evaluate our model and compare it with several state-of-the-art methods. Experimental results demonstrate that our proposed HyGNN model effectively predicts DDIs and impressively outperforms the baselines with a maximum ROC-AUC and PR-AUC of 97.9% and 98.1%, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge