Etienne Decencière
CMM
LmPT: Conditional Point Transformer for Anatomical Landmark Detection on 3D Point Clouds
Feb 02, 2026Abstract:Accurate identification of anatomical landmarks is crucial for various medical applications. Traditional manual landmarking is time-consuming and prone to inter-observer variability, while rule-based methods are often tailored to specific geometries or limited sets of landmarks. In recent years, anatomical surfaces have been effectively represented as point clouds, which are lightweight structures composed of spatial coordinates. Following this strategy and to overcome the limitations of existing landmarking techniques, we propose Landmark Point Transformer (LmPT), a method for automatic anatomical landmark detection on point clouds that can leverage homologous bones from different species for translational research. The LmPT model incorporates a conditioning mechanism that enables adaptability to different input types to conduct cross-species learning. We focus the evaluation of our approach on femoral landmarking using both human and newly annotated dog femurs, demonstrating its generalization and effectiveness across species. The code and dog femur dataset will be publicly available at: https://github.com/Pierreoo/LandmarkPointTransformer.
* This paper has been accepted at International Symposium on Biomedical Imaging (ISBI) 2026
Rethinking Metrics and Diffusion Architecture for 3D Point Cloud Generation
Nov 10, 2025Abstract:As 3D point clouds become a cornerstone of modern technology, the need for sophisticated generative models and reliable evaluation metrics has grown exponentially. In this work, we first expose that some commonly used metrics for evaluating generated point clouds, particularly those based on Chamfer Distance (CD), lack robustness against defects and fail to capture geometric fidelity and local shape consistency when used as quality indicators. We further show that introducing samples alignment prior to distance calculation and replacing CD with Density-Aware Chamfer Distance (DCD) are simple yet essential steps to ensure the consistency and robustness of point cloud generative model evaluation metrics. While existing metrics primarily focus on directly comparing 3D Euclidean coordinates, we present a novel metric, named Surface Normal Concordance (SNC), which approximates surface similarity by comparing estimated point normals. This new metric, when combined with traditional ones, provides a more comprehensive evaluation of the quality of generated samples. Finally, leveraging recent advancements in transformer-based models for point cloud analysis, such as serialized patch attention , we propose a new architecture for generating high-fidelity 3D structures, the Diffusion Point Transformer. We perform extensive experiments and comparisons on the ShapeNet dataset, showing that our model outperforms previous solutions, particularly in terms of quality of generated point clouds, achieving new state-of-the-art. Code available at https://github.com/matteo-bastico/DiffusionPointTransformer.
Euclidean Distance to Convex Polyhedra and Application to Class Representation in Spectral Images
Mar 26, 2025



Abstract:With the aim of estimating the abundance map from observations only, linear unmixing approaches are not always suitable to spectral images, especially when the number of bands is too small or when the spectra of the observed data are too correlated. To address this issue in the general case, we present a novel approach which provides an adapted spatial density function based on any arbitrary linear classifier. A robust mathematical formulation for computing the Euclidean distance to polyhedral sets is presented, along with an efficient algorithm that provides the exact minimum-norm point in a polyhedron. An empirical evaluation on the widely-used Samson hyperspectral dataset demonstrates that the proposed method surpasses state-of-the-art approaches in reconstructing abundance maps. Furthermore, its application to spectral images of a Lithium-ion battery, incompatible with linear unmixing models, validates the method's generality and effectiveness.
Coupled Laplacian Eigenmaps for Locally-Aware 3D Rigid Point Cloud Matching
Feb 27, 2024



Abstract:Point cloud matching, a crucial technique in computer vision, medical and robotics fields, is primarily concerned with finding correspondences between pairs of point clouds or voxels. In some practical scenarios, emphasizing local differences is crucial for accurately identifying a correct match, thereby enhancing the overall robustness and reliability of the matching process. Commonly used shape descriptors have several limitations and often fail to provide meaningful local insights on the paired geometries. In this work, we propose a new technique, based on graph Laplacian eigenmaps, to match point clouds by taking into account fine local structures. To deal with the order and sign ambiguity of Laplacian eigenmaps, we introduce a new operator, called Coupled Laplacian, that allows to easily generate aligned eigenspaces for multiple rigidly-registered geometries. We show that the similarity between those aligned high-dimensional spaces provides a locally meaningful score to match shapes. We initially evaluate the performance of the proposed technique in a point-wise manner, specifically focusing on the task of object anomaly localization using the MVTec 3D-AD dataset. Additionally, we define a new medical task, called automatic Bone Side Estimation (BSE), which we address through a global similarity score derived from coupled eigenspaces. In order to test it, we propose a benchmark collecting bone surface structures from various public datasets. Our matching technique, based on Coupled Laplacian, outperforms other methods by reaching an impressive accuracy on both tasks. The code to reproduce our experiments is publicly available at https://github.com/matteo-bastico/CoupledLaplacian and in the Supplementary Code.
A Simple and Robust Framework for Cross-Modality Medical Image Segmentation applied to Vision Transformers
Oct 09, 2023



Abstract:When it comes to clinical images, automatic segmentation has a wide variety of applications and a considerable diversity of input domains, such as different types of Magnetic Resonance Images (MRIs) and Computerized Tomography (CT) scans. This heterogeneity is a challenge for cross-modality algorithms that should equally perform independently of the input image type fed to them. Often, segmentation models are trained using a single modality, preventing generalization to other types of input data without resorting to transfer learning techniques. Furthermore, the multi-modal or cross-modality architectures proposed in the literature frequently require registered images, which are not easy to collect in clinical environments, or need additional processing steps, such as synthetic image generation. In this work, we propose a simple framework to achieve fair image segmentation of multiple modalities using a single conditional model that adapts its normalization layers based on the input type, trained with non-registered interleaved mixed data. We show that our framework outperforms other cross-modality segmentation methods, when applied to the same 3D UNet baseline model, on the Multi-Modality Whole Heart Segmentation Challenge. Furthermore, we define the Conditional Vision Transformer (C-ViT) encoder, based on the proposed cross-modality framework, and we show that it brings significant improvements to the resulting segmentation, up to 6.87\% of Dice accuracy, with respect to its baseline reference. The code to reproduce our experiments and the trained model weights are available at https://github.com/matteo-bastico/MI-Seg.
Heuristic Hyperparameter Choice for Image Anomaly Detection
Jul 20, 2023Abstract:Anomaly detection (AD) in images is a fundamental computer vision problem by deep learning neural network to identify images deviating significantly from normality. The deep features extracted from pretrained models have been proved to be essential for AD based on multivariate Gaussian distribution analysis. However, since models are usually pretrained on a large dataset for classification tasks such as ImageNet, they might produce lots of redundant features for AD, which increases computational cost and degrades the performance. We aim to do the dimension reduction of Negated Principal Component Analysis (NPCA) for these features. So we proposed some heuristic to choose hyperparameter of NPCA algorithm for getting as fewer components of features as possible while ensuring a good performance.
Adapting the Hypersphere Loss Function from Anomaly Detection to Anomaly Segmentation
Jan 23, 2023


Abstract:We propose an incremental improvement to Fully Convolutional Data Description (FCDD), an adaptation of the one-class classification approach from anomaly detection to image anomaly segmentation (a.k.a. anomaly localization). We analyze its original loss function and propose a substitute that better resembles its predecessor, the Hypersphere Classifier (HSC). Both are compared on the MVTec Anomaly Detection Dataset (MVTec-AD) -- training images are flawless objects/textures and the goal is to segment unseen defects -- showing that consistent improvement is achieved by better designing the pixel-wise supervision.
Giga-SSL: Self-Supervised Learning for Gigapixel Images
Dec 06, 2022



Abstract:Whole slide images (WSI) are microscopy images of stained tissue slides routinely prepared for diagnosis and treatment selection in medical practice. WSI are very large (gigapixel size) and complex (made of up to millions of cells). The current state-of-the-art (SoTA) approach to classify WSI subdivides them into tiles, encodes them by pre-trained networks and applies Multiple Instance Learning (MIL) to train for specific downstream tasks. However, annotated datasets are often small, typically a few hundred to a few thousand WSI, which may cause overfitting and underperforming models. Conversely, the number of unannotated WSI is ever increasing, with datasets of tens of thousands (soon to be millions) of images available. While it has been previously proposed to use these unannotated data to identify suitable tile representations by self-supervised learning (SSL), downstream classification tasks still require full supervision because parts of the MIL architecture is not trained during tile level SSL pre-training. Here, we propose a strategy of slide level SSL to leverage the large number of WSI without annotations to infer powerful slide representations. Applying our method to The Cancer-Genome Atlas, one of the most widely used data resources in cancer research (16 TB image data), we are able to downsize the dataset to 23 MB without any loss in predictive power: we show that a linear classifier trained on top of these embeddings maintains or improves previous SoTA performances on various benchmark WSI classification tasks. Finally, we observe that training a classifier on these representations with tiny datasets (e.g. 50 slides) improved performances over SoTA by an average of +6.3 AUC points over all downstream tasks.
[Reproducibility Report] Explainable Deep One-Class Classification
Jun 06, 2022![Figure 1 for [Reproducibility Report] Explainable Deep One-Class Classification](/_next/image?url=https%3A%2F%2Fai2-s2-public.s3.amazonaws.com%2Ffigures%2F2017-08-08%2Fc20a429ede0cb84dcece92c2346cffc906505256%2F3-Table1-1.png&w=640&q=75)
![Figure 2 for [Reproducibility Report] Explainable Deep One-Class Classification](/_next/image?url=https%3A%2F%2Fai2-s2-public.s3.amazonaws.com%2Ffigures%2F2017-08-08%2Fc20a429ede0cb84dcece92c2346cffc906505256%2F6-Figure1-1.png&w=640&q=75)
![Figure 3 for [Reproducibility Report] Explainable Deep One-Class Classification](/_next/image?url=https%3A%2F%2Fai2-s2-public.s3.amazonaws.com%2Ffigures%2F2017-08-08%2Fc20a429ede0cb84dcece92c2346cffc906505256%2F4-Table2-1.png&w=640&q=75)
![Figure 4 for [Reproducibility Report] Explainable Deep One-Class Classification](/_next/image?url=https%3A%2F%2Fai2-s2-public.s3.amazonaws.com%2Ffigures%2F2017-08-08%2Fc20a429ede0cb84dcece92c2346cffc906505256%2F6-Figure2-1.png&w=640&q=75)
Abstract:Fully Convolutional Data Description (FCDD), an explainable version of the Hypersphere Classifier (HSC), directly addresses image anomaly detection (AD) and pixel-wise AD without any post-hoc explainer methods. The authors claim that FCDD achieves results comparable with the state-of-the-art in sample-wise AD on Fashion-MNIST and CIFAR-10 and exceeds the state-of-the-art on the pixel-wise task on MVTec-AD. We reproduced the main results of the paper using the author's code with minor changes and provide runtime requirements to achieve if (CPU memory, GPU memory, and training time). We propose another analysis methodology using a critical difference diagram, and further investigate the test performance of the model during the training phase.
A modular U-Net for automated segmentation of X-ray tomography images in composite materials
Jul 15, 2021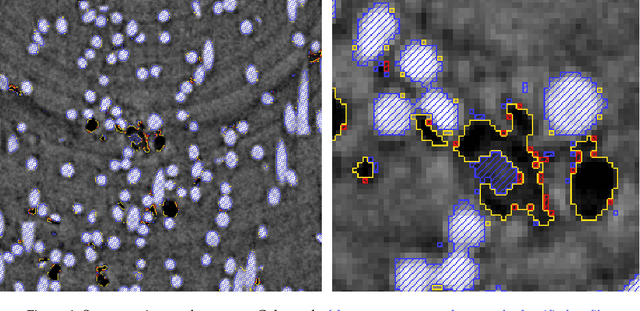
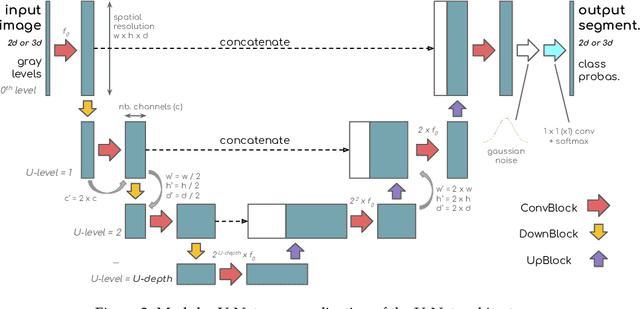
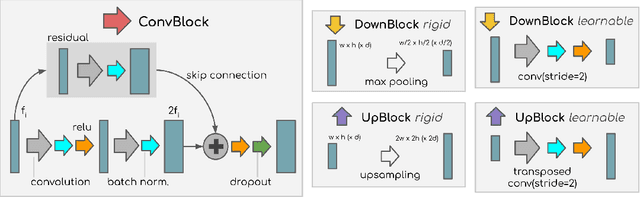
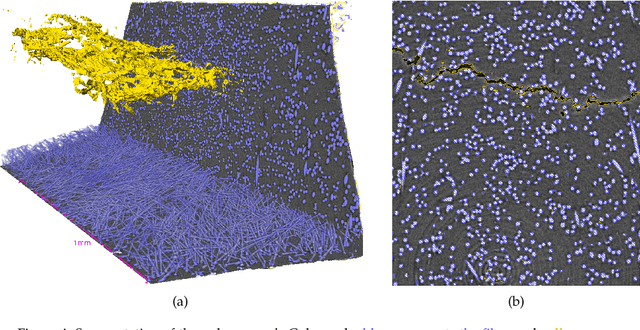
Abstract:X-ray Computed Tomography (XCT) techniques have evolved to a point that high-resolution data can be acquired so fast that classic segmentation methods are prohibitively cumbersome, demanding automated data pipelines capable of dealing with non-trivial 3D images. Deep learning has demonstrated success in many image processing tasks, including material science applications, showing a promising alternative for a humanfree segmentation pipeline. In this paper a modular interpretation of UNet (Modular U-Net) is proposed and trained to segment 3D tomography images of a three-phased glass fiber-reinforced Polyamide 66. We compare 2D and 3D versions of our model, finding that the former is slightly better than the latter. We observe that human-comparable results can be achievied even with only 10 annotated layers and using a shallow U-Net yields better results than a deeper one. As a consequence, Neural Network (NN) show indeed a promising venue to automate XCT data processing pipelines needing no human, adhoc intervention.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge