Dustin Scheinost
Unlearning Information Bottleneck: Machine Unlearning of Systematic Patterns and Biases
May 22, 2024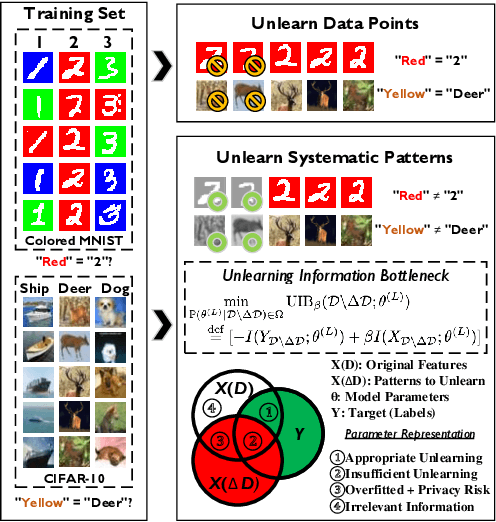
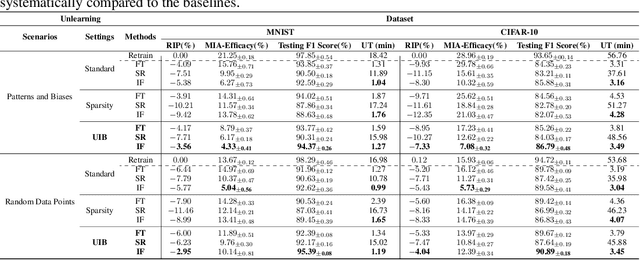
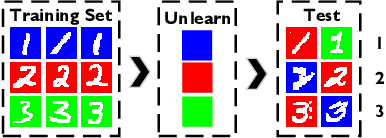
Abstract:Effective adaptation to distribution shifts in training data is pivotal for sustaining robustness in neural networks, especially when removing specific biases or outdated information, a process known as machine unlearning. Traditional approaches typically assume that data variations are random, which makes it difficult to adjust the model parameters accurately to remove patterns and characteristics from unlearned data. In this work, we present Unlearning Information Bottleneck (UIB), a novel information-theoretic framework designed to enhance the process of machine unlearning that effectively leverages the influence of systematic patterns and biases for parameter adjustment. By proposing a variational upper bound, we recalibrate the model parameters through a dynamic prior that integrates changes in data distribution with an affordable computational cost, allowing efficient and accurate removal of outdated or unwanted data patterns and biases. Our experiments across various datasets, models, and unlearning methods demonstrate that our approach effectively removes systematic patterns and biases while maintaining the performance of models post-unlearning.
Enhancement attacks in biomedical machine learning
Jan 05, 2023


Abstract:The prevalence of machine learning in biomedical research is rapidly growing, yet the trustworthiness of such research is often overlooked. While some previous works have investigated the ability of adversarial attacks to degrade model performance in medical imaging, the ability to falsely improve performance via recently-developed "enhancement attacks" may be a greater threat to biomedical machine learning. In the spirit of developing attacks to better understand trustworthiness, we developed three techniques to drastically enhance prediction performance of classifiers with minimal changes to features, including the enhancement of 1) within-dataset predictions, 2) a particular method over another, and 3) cross-dataset generalization. Our within-dataset enhancement framework falsely improved classifiers' accuracy from 50% to almost 100% while maintaining high feature similarities between original and enhanced data (Pearson's r's>0.99). Similarly, the method-specific enhancement framework was effective in falsely improving the performance of one method over another. For example, a simple neural network outperformed LR by 50% on our enhanced dataset, although no performance differences were present in the original dataset. Crucially, the original and enhanced data were still similar (r=0.95). Finally, we demonstrated that enhancement is not specific to within-dataset predictions but can also be adapted to enhance the generalization accuracy of one dataset to another by up to 38%. Overall, our results suggest that more robust data sharing and provenance tracking pipelines are necessary to maintain data integrity in biomedical machine learning research.
Data-driven mapping between functional connectomes using optimal transport
Jul 02, 2021



Abstract:Functional connectomes derived from functional magnetic resonance imaging have long been used to understand the functional organization of the brain. Nevertheless, a connectome is intrinsically linked to the atlas used to create it. In other words, a connectome generated from one atlas is different in scale and resolution compared to a connectome generated from another atlas. Being able to map connectomes and derived results between different atlases without additional pre-processing is a crucial step in improving interpretation and generalization between studies that use different atlases. Here, we use optimal transport, a powerful mathematical technique, to find an optimum mapping between two atlases. This mapping is then used to transform time series from one atlas to another in order to reconstruct a connectome. We validate our approach by comparing transformed connectomes against their "gold-standard" counterparts (i.e., connectomes generated directly from an atlas) and demonstrate the utility of transformed connectomes by applying these connectomes to predictive models based on a different atlas. We show that these transformed connectomes are significantly similar to their "gold-standard" counterparts and maintain individual differences in brain-behavior associations, demonstrating both the validity of our approach and its utility in downstream analyses. Overall, our approach is a promising avenue to increase the generalization of connectome-based results across different atlases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge