Dongyang Kuang
The Data Fusion Labeler (dFL): Challenges and Solutions to Data Harmonization, Labeling, and Provenance in Fusion Energy
Nov 12, 2025Abstract:Fusion energy research increasingly depends on the ability to integrate heterogeneous, multimodal datasets from high-resolution diagnostics, control systems, and multiscale simulations. The sheer volume and complexity of these datasets demand the development of new tools capable of systematically harmonizing and extracting knowledge across diverse modalities. The Data Fusion Labeler (dFL) is introduced as a unified workflow instrument that performs uncertainty-aware data harmonization, schema-compliant data fusion, and provenance-rich manual and automated labeling at scale. By embedding alignment, normalization, and labeling within a reproducible, operator-order-aware framework, dFL reduces time-to-analysis by greater than 50X (e.g., enabling >200 shots/hour to be consistently labeled rather than a handful per day), enhances label (and subsequently training) quality, and enables cross-device comparability. Case studies from DIII-D demonstrate its application to automated ELM detection and confinement regime classification, illustrating its potential as a core component of data-driven discovery, model validation, and real-time control in future burning plasma devices.
KAM -- a Kernel Attention Module for Emotion Classification with EEG Data
Aug 17, 2022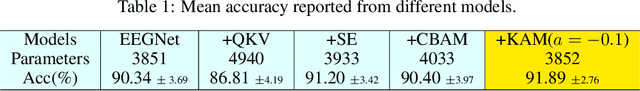
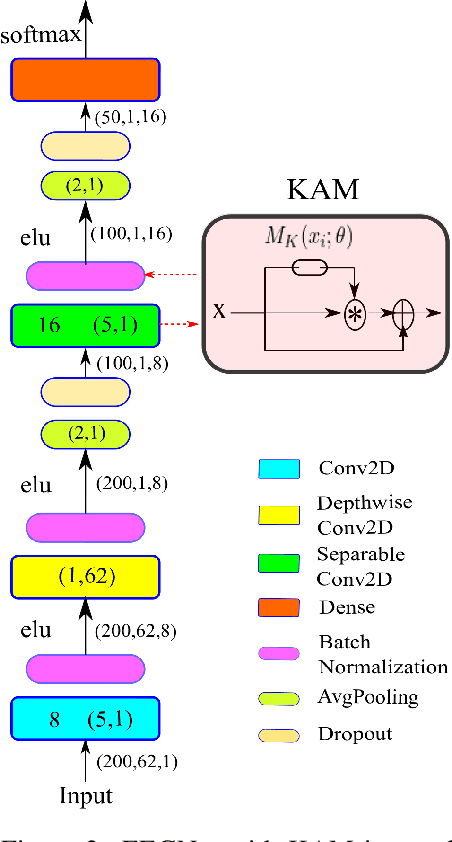
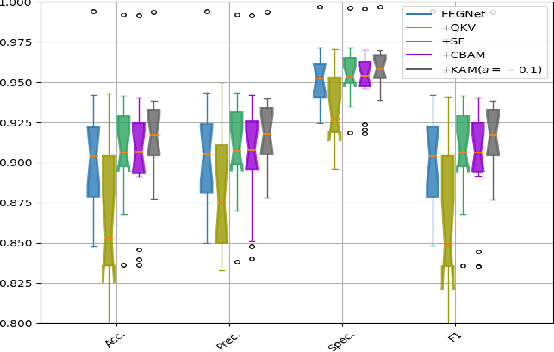
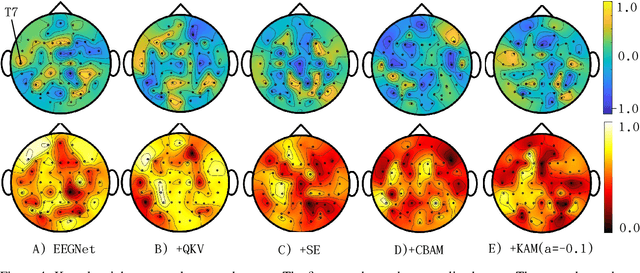
Abstract:In this work, a kernel attention module is presented for the task of EEG-based emotion classification with neural networks. The proposed module utilizes a self-attention mechanism by performing a kernel trick, demanding significantly fewer trainable parameters and computations than standard attention modules. The design also provides a scalar for quantitatively examining the amount of attention assigned during deep feature refinement, hence help better interpret a trained model. Using EEGNet as the backbone model, extensive experiments are conducted on the SEED dataset to assess the module's performance on within-subject classification tasks compared to other SOTA attention modules. Requiring only one extra parameter, the inserted module is shown to boost the base model's mean prediction accuracy up to more than 1\% across 15 subjects. A key component of the method is the interpretability of solutions, which is addressed using several different techniques, and is included throughout as part of the dependency analysis.
A Monotonicity Constrained Attention Module for Emotion Classification with Limited EEG Data
Aug 17, 2022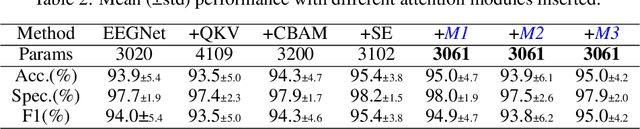
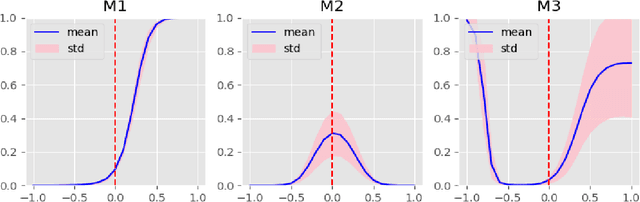
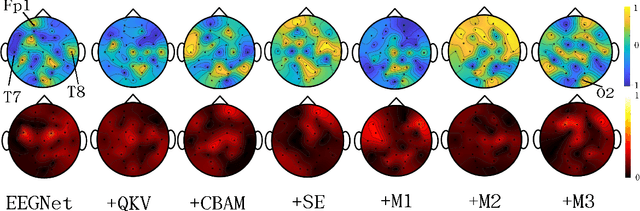
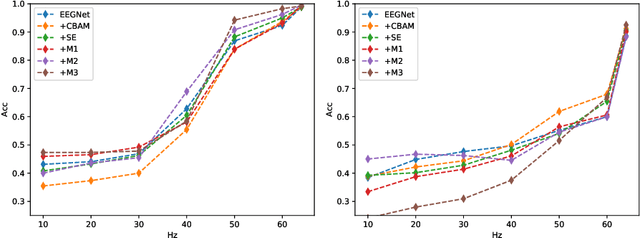
Abstract:In this work, a parameter-efficient attention module is presented for emotion classification using a limited, or relatively small, number of electroencephalogram (EEG) signals. This module is called the Monotonicity Constrained Attention Module (MCAM) due to its capability of incorporating priors on the monotonicity when converting features' Gram matrices into attention matrices for better feature refinement. Our experiments have shown that MCAM's effectiveness is comparable to state-of-the-art attention modules in boosting the backbone network's performance in prediction while requiring less parameters. Several accompanying sensitivity analyses on trained models' prediction concerning different attacks are also performed. These attacks include various frequency domain filtering levels and gradually morphing between samples associated with multiple labels. Our results can help better understand different modules' behaviour in prediction and can provide guidance in applications where data is limited and are with noises.
A 1d convolutional network for leaf and time series classification
Jun 28, 2019
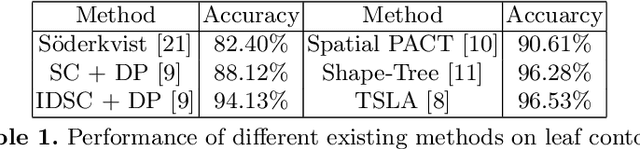
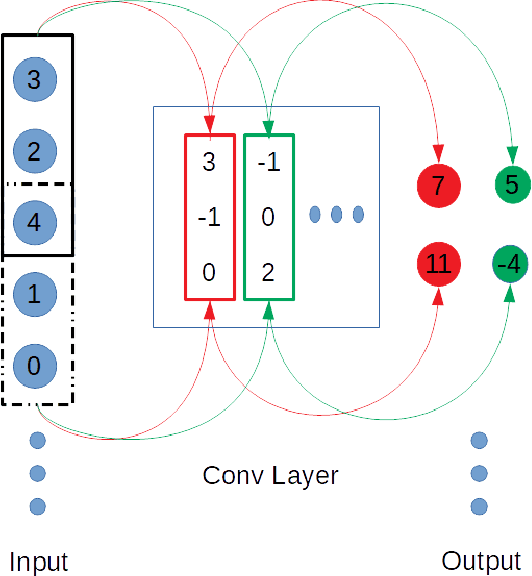

Abstract:In this paper, a 1d convolutional neural network is designed for classification tasks of leaves with centroid contour distance curve (CCDC) as the single feature. With this classifier, simple feature as CCDC shows more discriminating power than people thought previously. The same architecture can also be applied for classifying 1 dimensional time series with little changes. Experiments on some benchmark datasets shows this architecture can provide classification accuracies that are higher than some existing methods. Code for the paper is available at https://github.com/dykuang/Leaf Project.
On Reducing Negative Jacobian Determinant of the Deformation Predicted by Deep Registration Networks
Jun 28, 2019

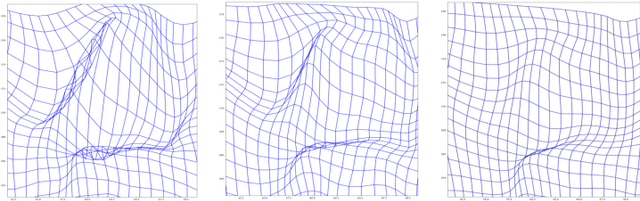
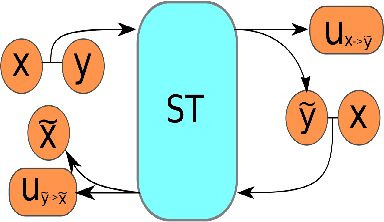
Abstract:Image registration is a fundamental step in medical image analysis. Ideally, the transformation that registers one image to another should be a diffeomorphism that is both invertible and smooth. Traditional methods like geodesic shooting approach the problem via differential geometry, with theoretical guarantees that the resulting transformation will be smooth and invertible. Most previous research using unsupervised deep neural networks for registration have used a local smoothness constraint (typically, a spatial variation loss) to address the smoothness issue. These networks usually produce non-invertible transformations with ``folding'' in multiple voxel locations, indicated by a negative determinant of the Jacobian matrix of the transformation. While using a loss function that specifically penalizes the folding is a straightforward solution, this usually requires carefully tuning the regularization strength, especially when there are also other losses. In this paper we address this problem from a different angle, by investigating possible training mechanisms that will help the network avoid negative Jacobians and produce smoother deformations. We contribute two independent ideas in this direction. Both ideas greatly reduce the number of folding locations in the predicted deformation, without making changes to the hyperparameters or the architecture used in the existing baseline registration network.
FAIM -- A ConvNet Method for Unsupervised 3D Medical Image Registration
Nov 22, 2018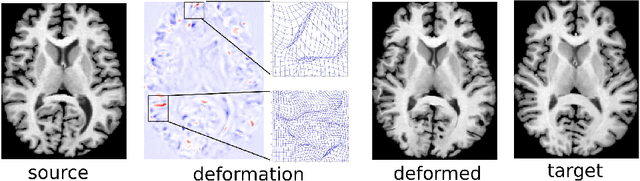
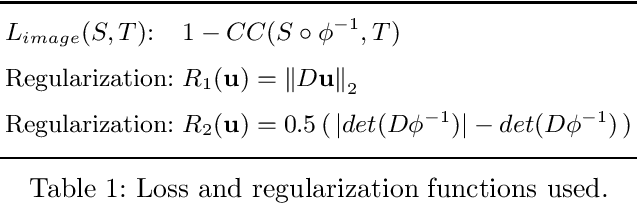
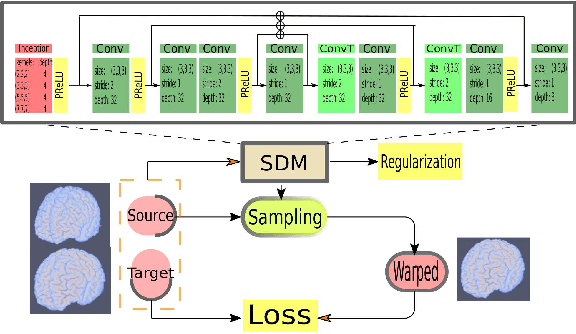

Abstract:We present a new unsupervised learning algorithm, "FAIM", for 3D medical image registration. Based on a convolutional neural net, FAIM learns from a training set of pairs of images, without needing ground truth information such as landmarks or dense registrations. Once trained, FAIM can register a new pair of images in less than a second, with competitive quality. We compared FAIM with a similar method, VoxelMorph, as well as a diffeomorphic method, uTIlzReg GeoShoot, on the LPBA40 and Mindboggle101 datasets. Results for FAIM were comparable or better than the other methods on pairwise registrations. The effect of different regularization choices on the predicted deformations is briefly investigated. Finally, an application to fast construction of a template and atlas is demonstrated.
A landmark-based algorithm for automatic pattern recognition and abnormality detection
May 11, 2017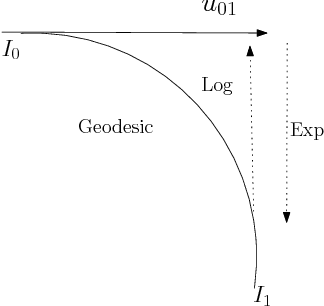
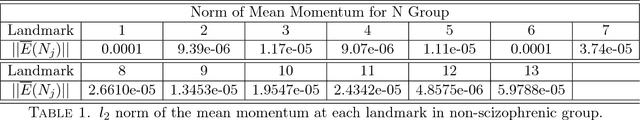
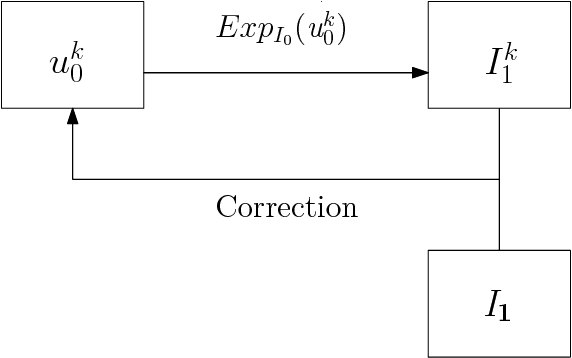
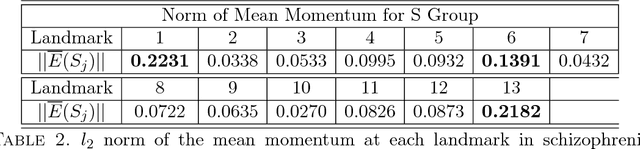
Abstract:We study a class of mathematical and statistical algorithms with the aim of establishing a computer-based framework for fast and reliable automatic abnormality detection on landmark represented image templates. Under this framework, we apply a landmark-based algorithm for finding a group average as an estimator that is said to best represent the common features of the group in study. This algorithm extracts information of momentum at each landmark through the process of template matching. If ever converges, the proposed algorithm produces a local coordinate system for each member of the observing group, in terms of the residual momentum. We use a Bayesian approach on the collected residual momentum representations for making inference. For illustration, we apply this framework to a small database of brain images for detecting structure abnormality. The brain structure changes identified by our framework are highly consistent with studies in the literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge