Dominik Drees
A Bhattacharyya Coefficient-Based Framework for Noise Model-Aware Random Walker Image Segmentation
Jun 02, 2022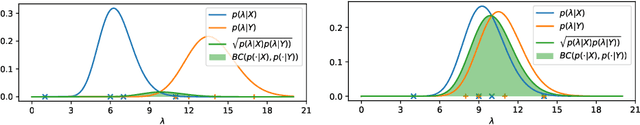
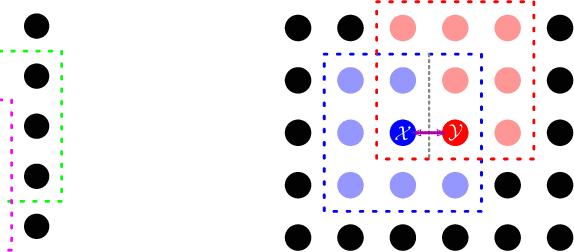
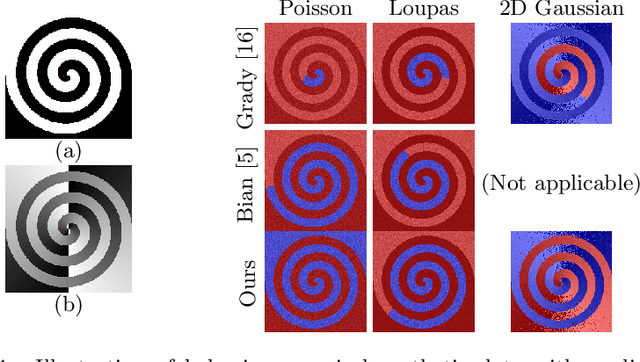
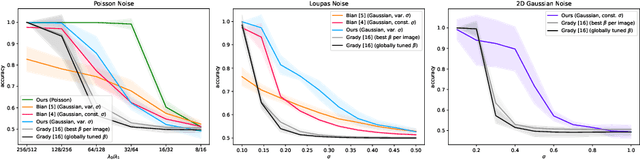
Abstract:One well established method of interactive image segmentation is the random walker algorithm. Considerable research on this family of segmentation methods has been continuously conducted in recent years with numerous applications. These methods are common in using a simple Gaussian weight function which depends on a parameter that strongly influences the segmentation performance. In this work we propose a general framework of deriving weight functions based on probabilistic modeling. This framework can be concretized to cope with virtually any well-defined noise model. It eliminates the critical parameter and thus avoids time-consuming parameter search. We derive the specific weight functions for common noise types and show their superior performance on synthetic data as well as different biomedical image data (MRI images from the NYU fastMRI dataset, larvae images acquired with the FIM technique). Our framework can also be used in multiple other applications, e.g., the graph cut algorithm and its extensions.
A Streaming Volumetric Image Generation Framework for Development and Evaluation of Out-of-Core Methods
Dec 18, 2021



Abstract:Advances in 3D imaging technology in recent years have allowed for increasingly high resolution volumetric images of large specimen. The resulting datasets of hundreds of Gigabytes in size call for new scalable and memory efficient approaches in the field of image processing, where some progress has been made already. At the same time, quantitative evaluation of these new methods is difficult both in terms of the availability of specific data sizes and in the generation of associated ground truth data. In this paper we present an algorithmic framework that can be used to efficiently generate test (and ground truth) volume data, optionally even in a streaming fashion. As the proposed nested sweeps algorithm is fast, it can be used to generate test data on demand. We analyze the asymptotic run time of the presented algorithm and compare it experimentally to alternative approaches as well as a hypothetical best-case baseline method. In a case study, the framework is applied to the popular VascuSynth software for vascular image generation, making it capable of efficiently producing larger-than-main memory volumes which is demonstrated by generating a trillion voxel (1TB) image. Implementations of the presented framework are available online in the form of the modified version of Vascusynth and the code used for the experimental evaluation. In addition, the test data generation procedure has been integrated into the popular volume rendering and processing framework Voreen.
Hierarchical Random Walker Segmentation for Large Volumetric Biomedical Data
Mar 17, 2021



Abstract:The random walker method for image segmentation is a popular tool for semi-automatic image segmentation, especially in the biomedical field. However, its linear asymptotic run time and memory requirements make application to 3D datasets of increasing sizes impractical. We propose a hierarchical framework that, to the best of our knowledge, is the first attempt to overcome these restrictions for the random walker algorithm and achieves sublinear run time and constant memory complexity. The method is evaluated on synthetic data and real data from current biomedical research, where high segmentation quality is quantitatively confirmed and visually observed, respectively. The incremental (i.e., interaction update) run time is demonstrated to be in seconds on a standard PC even for volumes of hundreds of Gigabytes in size. An implementation of the presented method is publicly available in version 5.2 of the widely used volume rendering and processing software Voreen (https://www.uni-muenster.de/Voreen/).
Scalable Robust Graph and Feature Extraction for Arbitrary Vessel Networks in Large Volumetric Datasets
Feb 05, 2021



Abstract:Recent advances in 3D imaging technologies provide novel insights to researchers and reveal finer and more detail of examined specimen, especially in the biomedical domain, but also impose huge challenges regarding scalability for automated analysis algorithms due to rapidly increasing dataset sizes. In particular, existing research towards automated vessel network analysis does not consider memory requirements of proposed algorithms and often generates a large number of spurious branches for structures consisting of many voxels. Additionally, very often these algorithms have further restrictions such as the limitation to tree topologies or relying on the properties of specific image modalities. We present a scalable pipeline (in terms of computational cost, required main memory and robustness) that extracts an annotated abstract graph representation from the foreground segmentation of vessel networks of arbitrary topology and vessel shape. Only a single, dimensionless, a-priori determinable parameter is required. By careful engineering of individual pipeline stages and a novel iterative refinement scheme we are, for the first time, able to analyze the topology of volumes of roughly 1TB on commodity hardware. An implementation of the presented pipeline is publicly available in version 5.1 of the volume rendering and processing engine Voreen (https://www.uni-muenster.de/Voreen/).
Barista - a Graphical Tool for Designing and Training Deep Neural Networks
Feb 13, 2018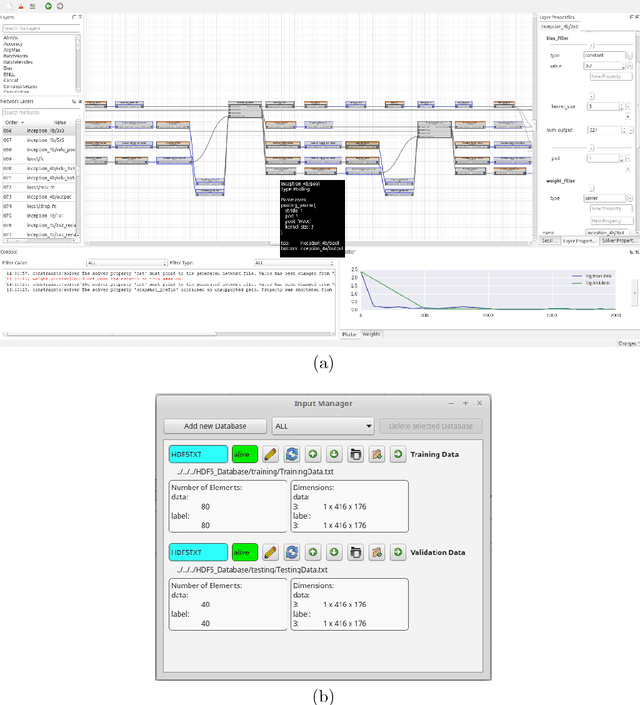
Abstract:In recent years, the importance of deep learning has significantly increased in pattern recognition, computer vision, and artificial intelligence research, as well as in industry. However, despite the existence of multiple deep learning frameworks, there is a lack of comprehensible and easy-to-use high-level tools for the design, training, and testing of deep neural networks (DNNs). In this paper, we introduce Barista, an open-source graphical high-level interface for the Caffe deep learning framework. While Caffe is one of the most popular frameworks for training DNNs, editing prototext files in order to specify the net architecture and hyper parameters can become a cumbersome and error-prone task. Instead, Barista offers a fully graphical user interface with a graph-based net topology editor and provides an end-to-end training facility for DNNs, which allows researchers to focus on solving their problems without having to write code, edit text files, or manually parse logged data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge