Friedemann Kiefer
Active Axial Motion Compensation in Multiphoton-Excited Fluorescence Microscopy
Jan 19, 2024

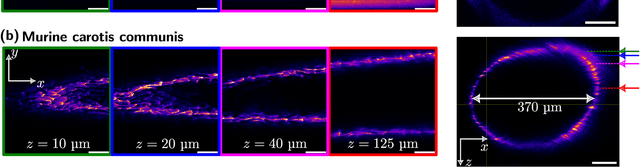

Abstract:In living organisms, the natural motion caused by the heartbeat, breathing, or muscle movements leads to the deformation of tissue caused by translation and stretching of the tissue structure. This effect results in the displacement or deformation of the plane of observation for intravital microscopy and causes motion-induced aberrations of the resulting image data. This, in turn, places severe limitations on the time during which specific events can be observed in intravital imaging experiments. These limitations can be overcome if the tissue motion can be compensated such that the plane of observation remains steady. We have developed a mathematical shape space model that can predict the periodic motion of a cylindrical tissue phantom resembling blood vessels. This model is then used to rapidly calculate the future position of the plane of observation of a confocal multiphoton fluorescence microscope. The focal plane is continuously adjusted to the calculated position with a piezo-actuated objective lens holder. We demonstrate active motion compensation for non-harmonic axial displacements of the vessel phantom with a field of view up to 400 $\mu$m $\times$ 400 $\mu$m, vertical amplitudes of more than 100 $\mu$m, and at a rate of 0.5 Hz.
Scalable Robust Graph and Feature Extraction for Arbitrary Vessel Networks in Large Volumetric Datasets
Feb 05, 2021



Abstract:Recent advances in 3D imaging technologies provide novel insights to researchers and reveal finer and more detail of examined specimen, especially in the biomedical domain, but also impose huge challenges regarding scalability for automated analysis algorithms due to rapidly increasing dataset sizes. In particular, existing research towards automated vessel network analysis does not consider memory requirements of proposed algorithms and often generates a large number of spurious branches for structures consisting of many voxels. Additionally, very often these algorithms have further restrictions such as the limitation to tree topologies or relying on the properties of specific image modalities. We present a scalable pipeline (in terms of computational cost, required main memory and robustness) that extracts an annotated abstract graph representation from the foreground segmentation of vessel networks of arbitrary topology and vessel shape. Only a single, dimensionless, a-priori determinable parameter is required. By careful engineering of individual pipeline stages and a novel iterative refinement scheme we are, for the first time, able to analyze the topology of volumes of roughly 1TB on commodity hardware. An implementation of the presented pipeline is publicly available in version 5.1 of the volume rendering and processing engine Voreen (https://www.uni-muenster.de/Voreen/).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge