Dmitry A. Konovalov
Mars Spectrometry 2: Gas Chromatography -- Second place solution
Mar 24, 2024Abstract:The Mars Spectrometry 2: Gas Chromatography challenge was sponsored by NASA and run on the DrivenData competition platform in 2022. This report describes the solution which achieved the second-best score on the competition's test dataset. The solution utilized two-dimensional, image-like representations of the competition's chromatography data samples. A number of different Convolutional Neural Network models were trained and ensembled for the final submission.
A Realistic Fish-Habitat Dataset to Evaluate Algorithms for Underwater Visual Analysis
Aug 28, 2020

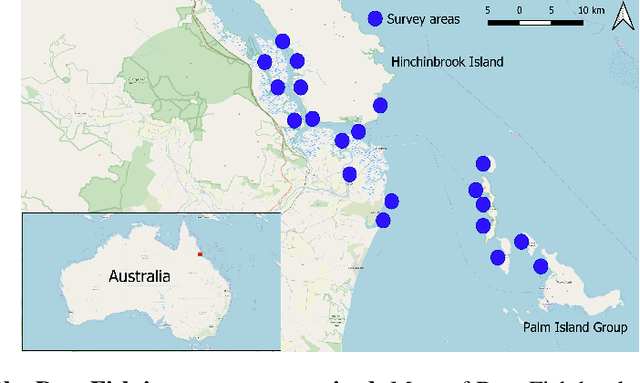

Abstract:Visual analysis of complex fish habitats is an important step towards sustainable fisheries for human consumption and environmental protection. Deep Learning methods have shown great promise for scene analysis when trained on large-scale datasets. However, current datasets for fish analysis tend to focus on the classification task within constrained, plain environments which do not capture the complexity of underwater fish habitats. To address this limitation, we present DeepFish as a benchmark suite with a large-scale dataset to train and test methods for several computer vision tasks. The dataset consists of approximately 40 thousand images collected underwater from 20 \green{habitats in the} marine-environments of tropical Australia. The dataset originally contained only classification labels. Thus, we collected point-level and segmentation labels to have a more comprehensive fish analysis benchmark. These labels enable models to learn to automatically monitor fish count, identify their locations, and estimate their sizes. Our experiments provide an in-depth analysis of the dataset characteristics, and the performance evaluation of several state-of-the-art approaches based on our benchmark. Although models pre-trained on ImageNet have successfully performed on this benchmark, there is still room for improvement. Therefore, this benchmark serves as a testbed to motivate further development in this challenging domain of underwater computer vision. Code is available at: https://github.com/alzayats/DeepFish
Data-Efficient Classification of Birdcall Through Convolutional Neural Networks Transfer Learning
Sep 17, 2019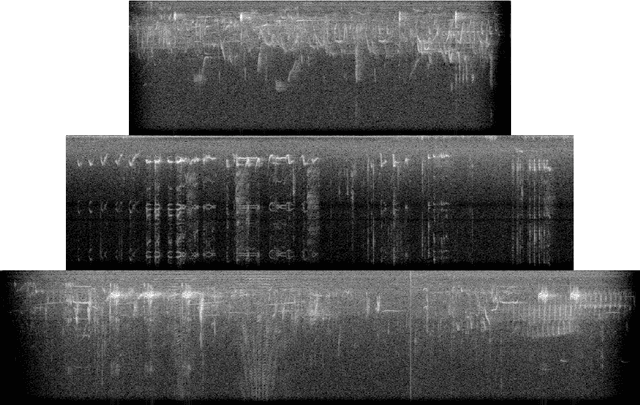
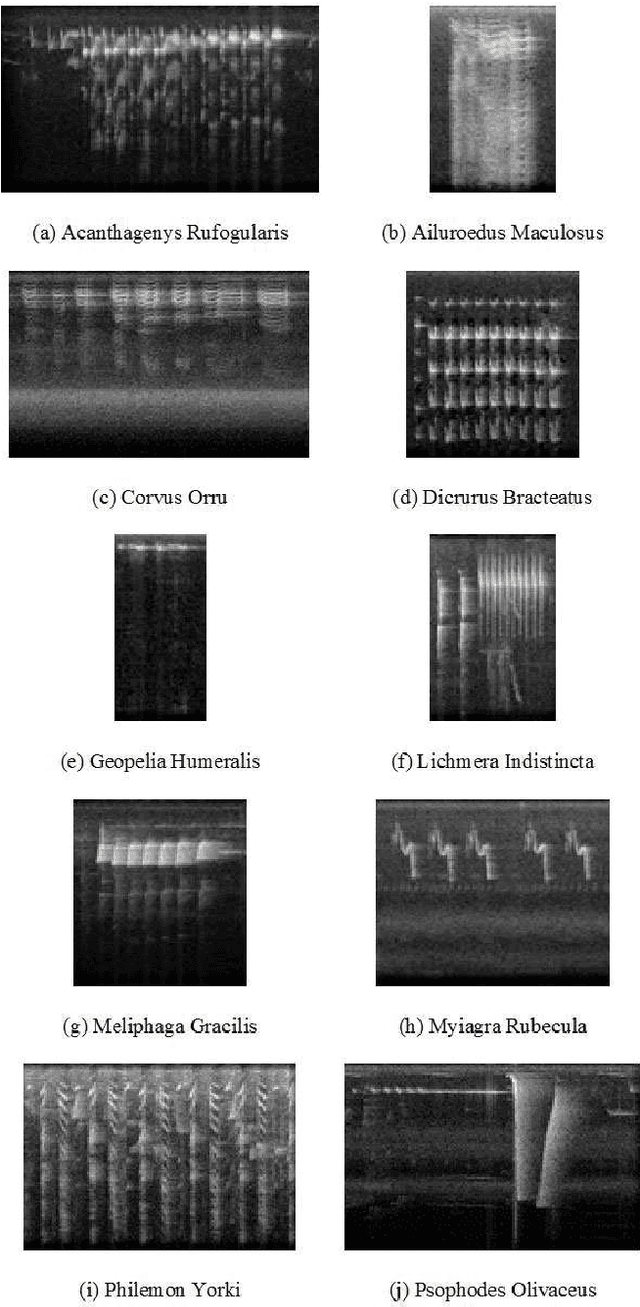
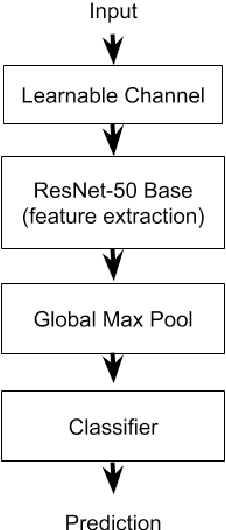
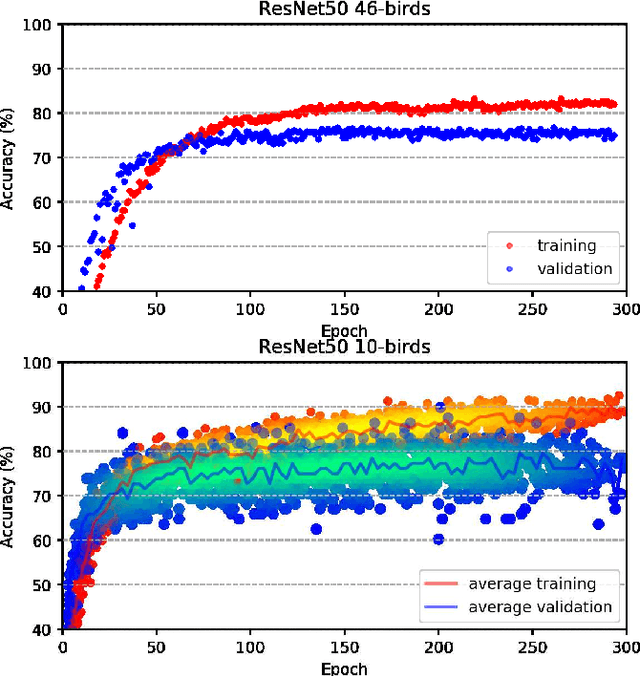
Abstract:Deep learning Convolutional Neural Network (CNN) models are powerful classification models but require a large amount of training data. In niche domains such as bird acoustics, it is expensive and difficult to obtain a large number of training samples. One method of classifying data with a limited number of training samples is to employ transfer learning. In this research, we evaluated the effectiveness of birdcall classification using transfer learning from a larger base dataset (2814 samples in 46 classes) to a smaller target dataset (351 samples in 10 classes) using the ResNet-50 CNN. We obtained 79% average validation accuracy on the target dataset in 5-fold cross-validation. The methodology of transfer learning from an ImageNet-trained CNN to a project-specific and a much smaller set of classes and images was extended to the domain of spectrogram images, where the base dataset effectively played the role of the ImageNet.
Automatic segmentation of kidney and liver tumors in CT images
Sep 16, 2019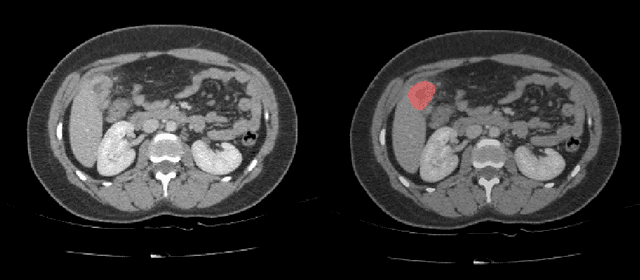
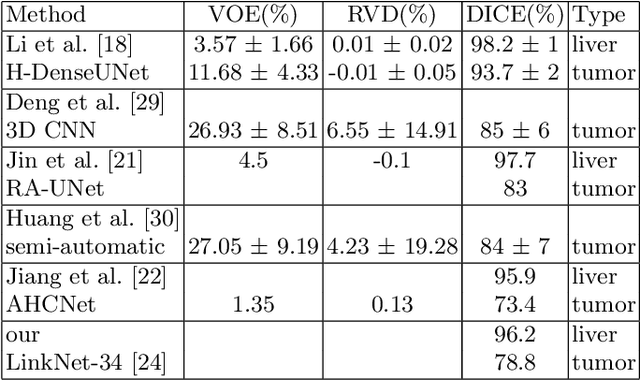
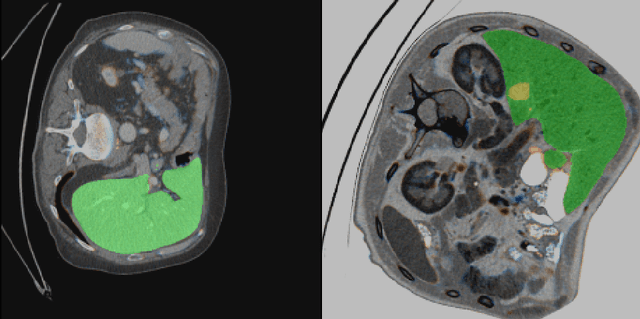
Abstract:Automatic segmentation of hepatic lesions in computed tomography (CT) images is a challenging task to perform due to heterogeneous, diffusive shape of tumors and complex background. To address the problem more and more researchers rely on assistance of deep convolutional neural networks (CNN) with 2D or 3D type architecture that have proven to be effective in a wide range of computer vision tasks, including medical image processing. In this technical report, we carry out research focused on more careful approach to the process of learning rather than on complex architecture of the CNN. We have chosen MICCAI 2017 LiTS dataset for training process and the public 3DIRCADb dataset for validation of our method. The proposed algorithm reached DICE score 78.8% on the 3DIRCADb dataset. The described method was then applied to the 2019 Kidney Tumor Segmentation (KiTS-2019) challenge, where our single submission achieved 96.38% for kidney and 67.38% for tumor Dice scores.
Automatic Weight Estimation of Harvested Fish from Images
Sep 06, 2019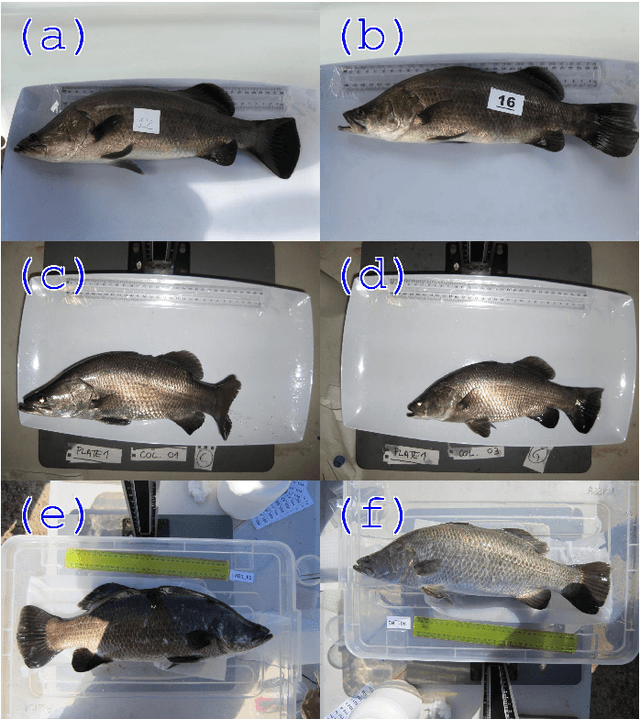

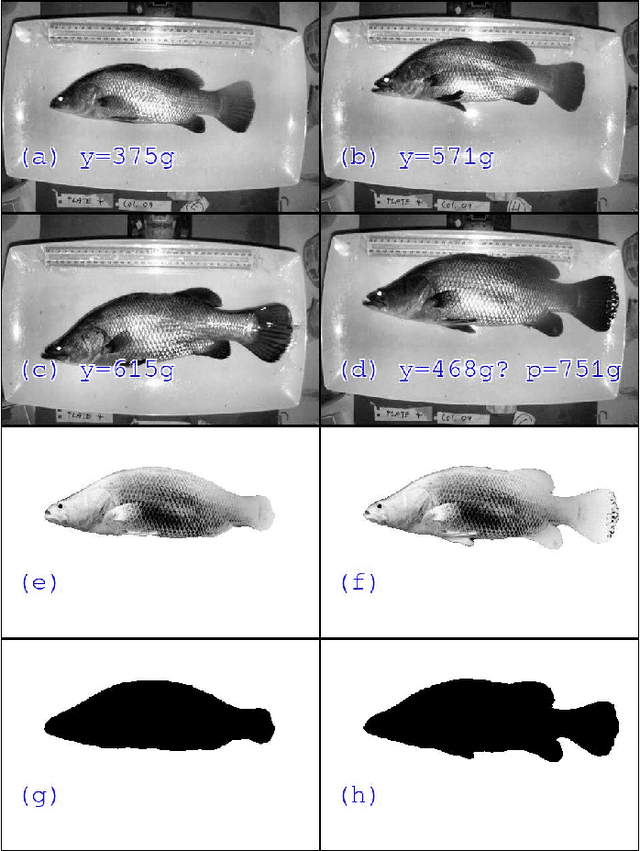
Abstract:Approximately 2,500 weights and corresponding images of harvested Lates calcarifer (Asian seabass or barramundi) were collected at three different locations in Queensland, Australia. Two instances of the LinkNet-34 segmentation Convolutional Neural Network (CNN) were trained. The first one was trained on 200 manually segmented fish masks with excluded fins and tails. The second was trained on 100 whole-fish masks. The two CNNs were applied to the rest of the images and yielded automatically segmented masks. The one-factor and two-factor simple mathematical weight-from-area models were fitted on 1072 area-weight pairs from the first two locations, where area values were extracted from the automatically segmented masks. When applied to 1,400 test images (from the third location), the one-factor whole-fish mask model achieved the best mean absolute percentage error (MAPE), MAPE=4.36%. Direct weight-from-image regression CNNs were also trained, where the no-fins based CNN performed best on the test images with MAPE=4.28%.
In Situ Cane Toad Recognition
Jun 09, 2019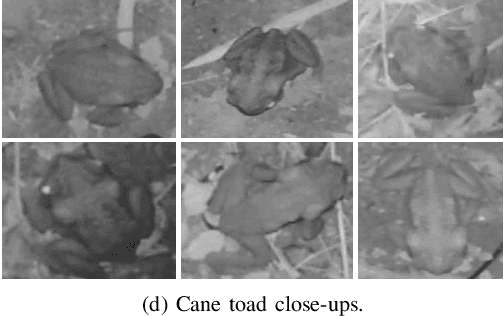
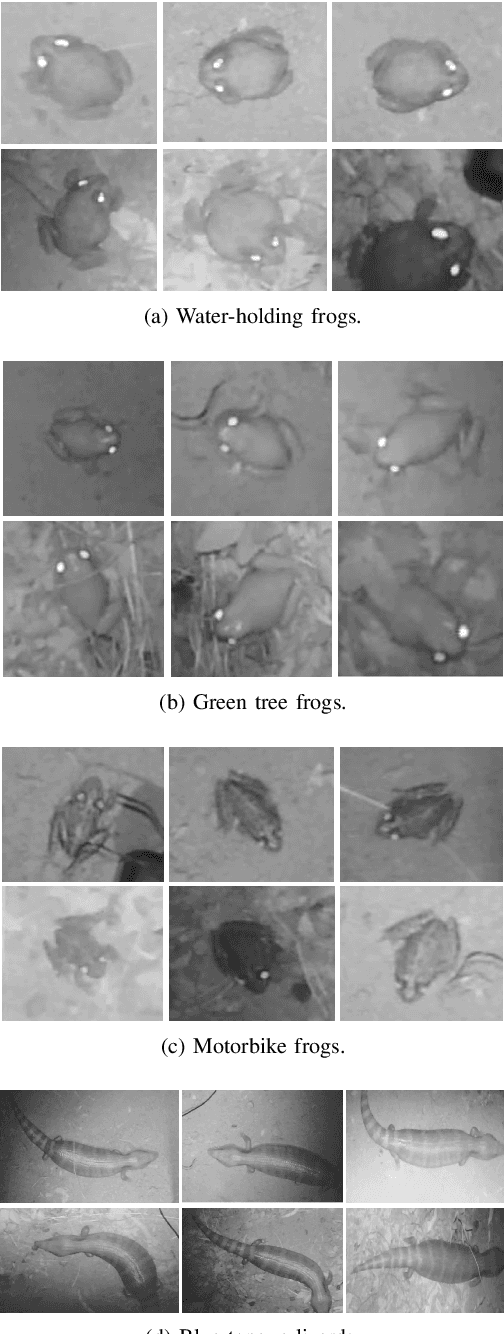


Abstract:Cane toads are invasive, toxic to native predators, compete with native insectivores, and have a devastating impact on Australian ecosystems, prompting the Australian government to list toads as a key threatening process under the Environment Protection and Biodiversity Conservation Act 1999. Mechanical cane toad traps could be made more native-fauna friendly if they could distinguish invasive cane toads from native species. Here we designed and trained a Convolution Neural Network (CNN) starting from the Xception CNN. The XToadGmp toad-recognition CNN we developed was trained end-to-end using heat-map Gaussian targets. After training, XToadGmp required minimum image pre/post-processing and when tested on 720x1280 shaped images, it achieved 97.1% classification accuracy on 1863 toad and 2892 not-toad test images, which were not used in training.
* Accepted for DICTA2018 https://doi.org/10.1109/DICTA.2018.8615780
Underwater Fish Detection with Weak Multi-Domain Supervision
May 26, 2019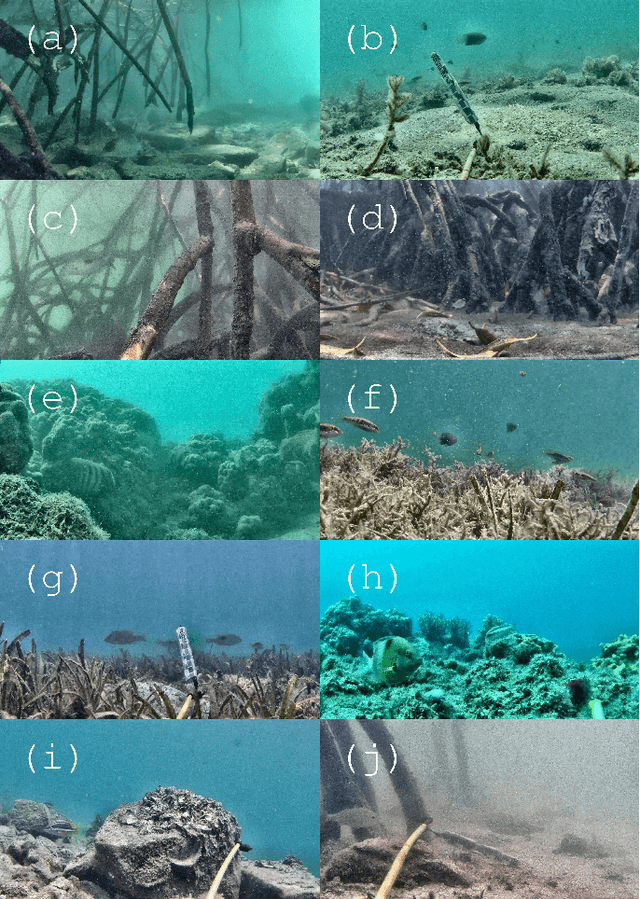
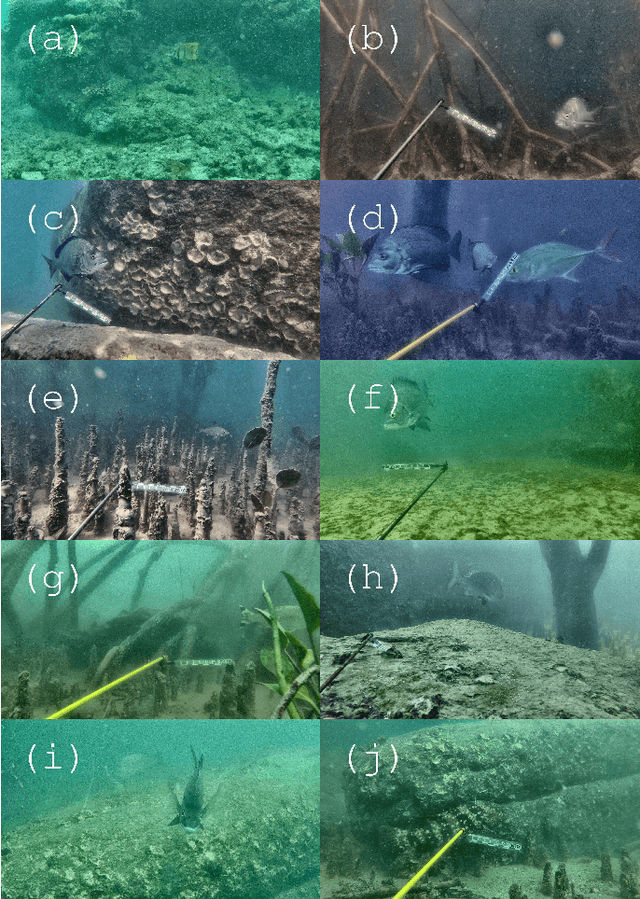


Abstract:Given a sufficiently large training dataset, it is relatively easy to train a modern convolution neural network (CNN) as a required image classifier. However, for the task of fish classification and/or fish detection, if a CNN was trained to detect or classify particular fish species in particular background habitats, the same CNN exhibits much lower accuracy when applied to new/unseen fish species and/or fish habitats. Therefore, in practice, the CNN needs to be continuously fine-tuned to improve its classification accuracy to handle new project-specific fish species or habitats. In this work we present a labelling-efficient method of training a CNN-based fish-detector (the Xception CNN was used as the base) on relatively small numbers (4,000) of project-domain underwater fish/no-fish images from 20 different habitats. Additionally, 17,000 of known negative (that is, missing fish) general-domain (VOC2012) above-water images were used. Two publicly available fish-domain datasets supplied additional 27,000 of above-water and underwater positive/fish images. By using this multi-domain collection of images, the trained Xception-based binary (fish/not-fish) classifier achieved 0.17% false-positives and 0.61% false-negatives on the project's 20,000 negative and 16,000 positive holdout test images, respectively. The area under the ROC curve (AUC) was 99.94%.
DeepWeeds: A Multiclass Weed Species Image Dataset for Deep Learning
Oct 09, 2018



Abstract:Robotic weed control has seen increased research in the past decade with its potential for boosting productivity in agriculture. Majority of works focus on developing robotics for arable croplands, ignoring the significant weed management problems facing rangeland stock farmers. Perhaps the greatest obstacle to widespread uptake of robotic weed control is the robust detection of weed species in their natural environment. The unparalleled successes of deep learning make it an ideal candidate for recognising various weed species in the highly complex Australian rangeland environment. This work contributes the first large, public, multiclass image dataset of weed species from the Australian rangelands; allowing for the development of robust detection methods to make robotic weed control viable. The DeepWeeds dataset consists of 17,509 labelled images of eight nationally significant weed species native to eight locations across northern Australia. This paper also presents a baseline for classification performance on the dataset using the benchmark deep learning models, Inception-v3 and ResNet-50. These models achieved an average classification performance of 87.9% and 90.5%, respectively. This strong result bodes well for future field implementation of robotic weed control methods in the Australian rangelands.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge