Dina B. Efremova
Data-Efficient Classification of Birdcall Through Convolutional Neural Networks Transfer Learning
Sep 17, 2019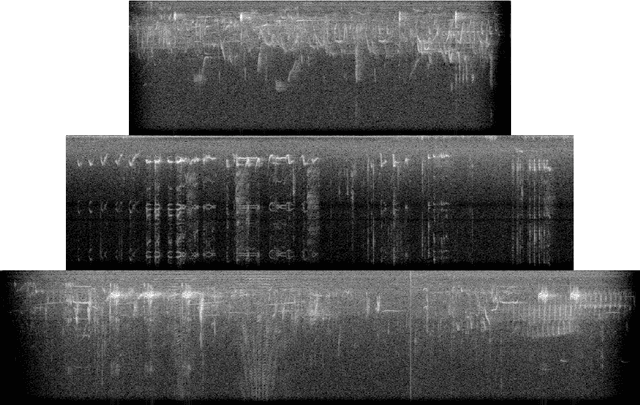
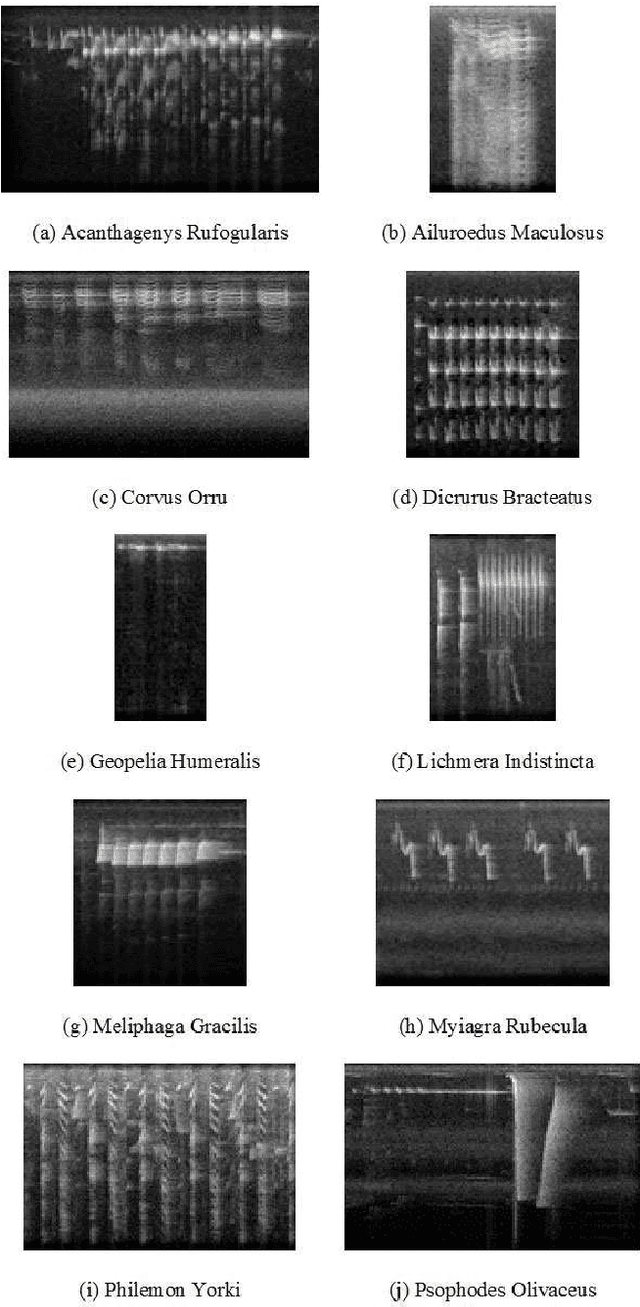
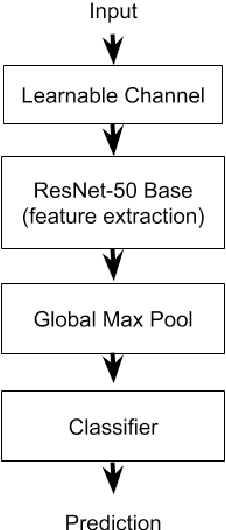
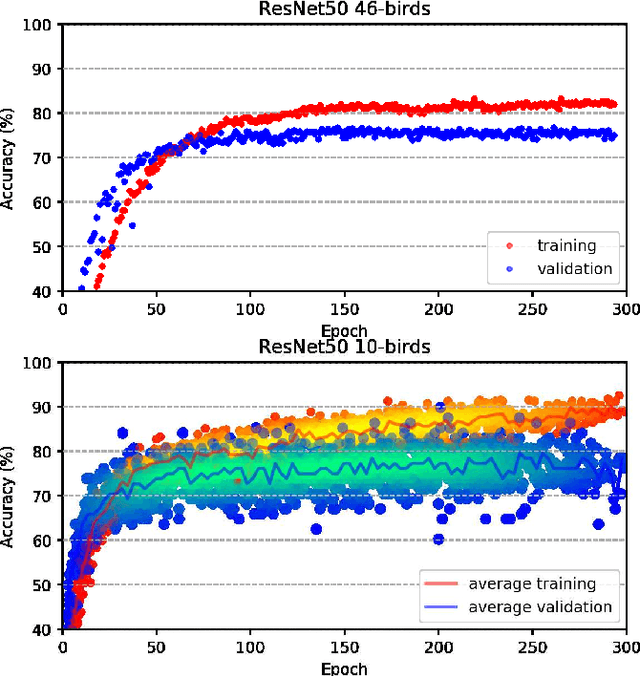
Abstract:Deep learning Convolutional Neural Network (CNN) models are powerful classification models but require a large amount of training data. In niche domains such as bird acoustics, it is expensive and difficult to obtain a large number of training samples. One method of classifying data with a limited number of training samples is to employ transfer learning. In this research, we evaluated the effectiveness of birdcall classification using transfer learning from a larger base dataset (2814 samples in 46 classes) to a smaller target dataset (351 samples in 10 classes) using the ResNet-50 CNN. We obtained 79% average validation accuracy on the target dataset in 5-fold cross-validation. The methodology of transfer learning from an ImageNet-trained CNN to a project-specific and a much smaller set of classes and images was extended to the domain of spectrogram images, where the base dataset effectively played the role of the ImageNet.
Automatic segmentation of kidney and liver tumors in CT images
Sep 16, 2019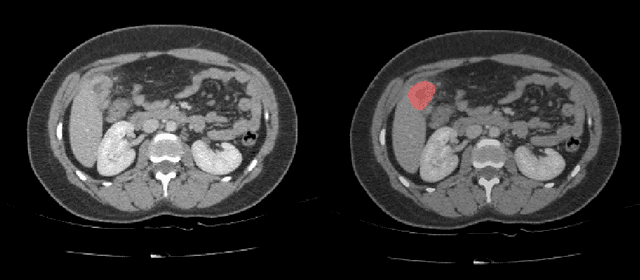
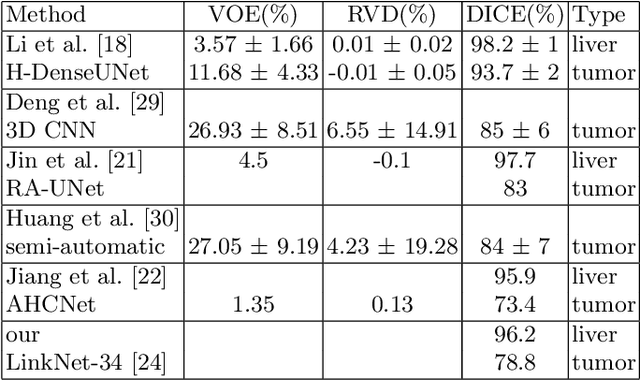
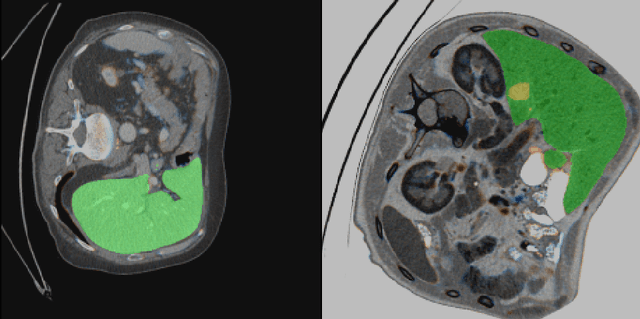
Abstract:Automatic segmentation of hepatic lesions in computed tomography (CT) images is a challenging task to perform due to heterogeneous, diffusive shape of tumors and complex background. To address the problem more and more researchers rely on assistance of deep convolutional neural networks (CNN) with 2D or 3D type architecture that have proven to be effective in a wide range of computer vision tasks, including medical image processing. In this technical report, we carry out research focused on more careful approach to the process of learning rather than on complex architecture of the CNN. We have chosen MICCAI 2017 LiTS dataset for training process and the public 3DIRCADb dataset for validation of our method. The proposed algorithm reached DICE score 78.8% on the 3DIRCADb dataset. The described method was then applied to the 2019 Kidney Tumor Segmentation (KiTS-2019) challenge, where our single submission achieved 96.38% for kidney and 67.38% for tumor Dice scores.
Automatic Weight Estimation of Harvested Fish from Images
Sep 06, 2019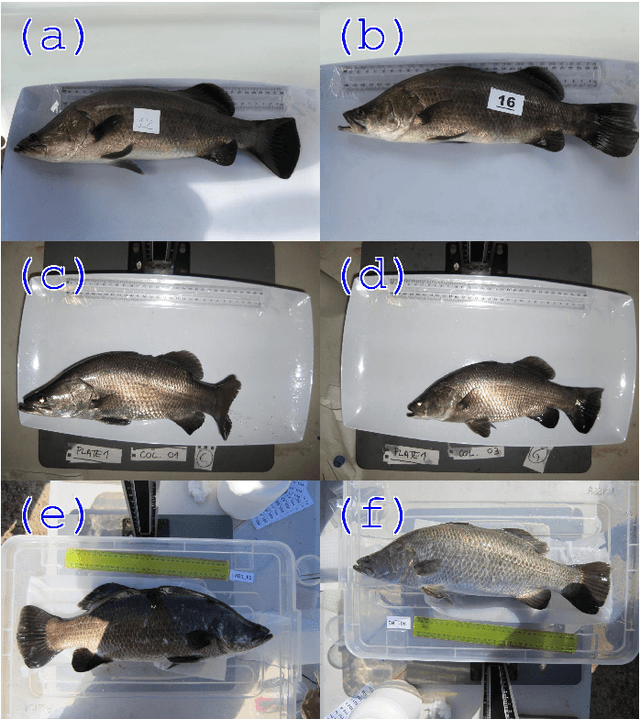

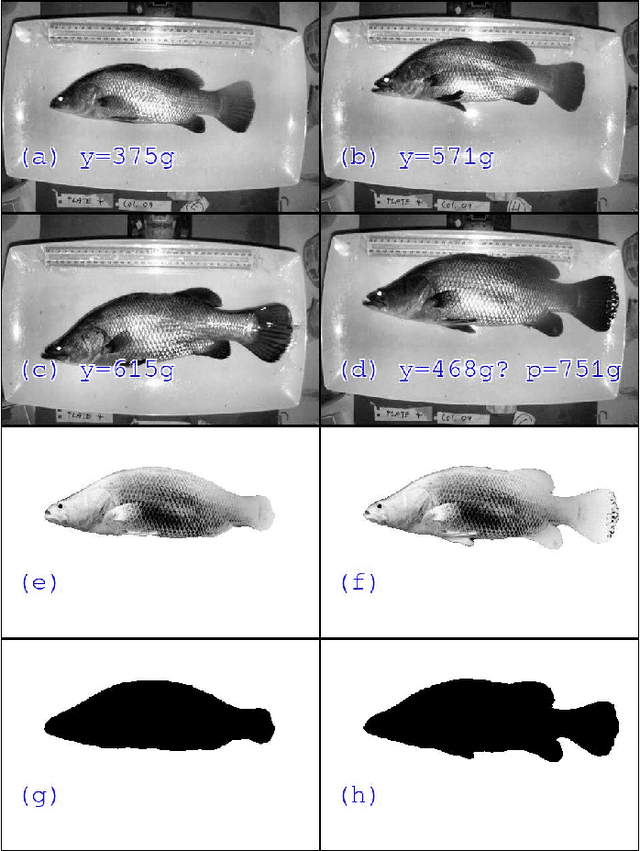
Abstract:Approximately 2,500 weights and corresponding images of harvested Lates calcarifer (Asian seabass or barramundi) were collected at three different locations in Queensland, Australia. Two instances of the LinkNet-34 segmentation Convolutional Neural Network (CNN) were trained. The first one was trained on 200 manually segmented fish masks with excluded fins and tails. The second was trained on 100 whole-fish masks. The two CNNs were applied to the rest of the images and yielded automatically segmented masks. The one-factor and two-factor simple mathematical weight-from-area models were fitted on 1072 area-weight pairs from the first two locations, where area values were extracted from the automatically segmented masks. When applied to 1,400 test images (from the third location), the one-factor whole-fish mask model achieved the best mean absolute percentage error (MAPE), MAPE=4.36%. Direct weight-from-image regression CNNs were also trained, where the no-fins based CNN performed best on the test images with MAPE=4.28%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge