Dipak K. Dey
Distribution-free inference for LightGBM and GLM with Tweedie loss
Jul 09, 2025Abstract:Prediction uncertainty quantification is a key research topic in recent years scientific and business problems. In insurance industries (\cite{parodi2023pricing}), assessing the range of possible claim costs for individual drivers improves premium pricing accuracy. It also enables insurers to manage risk more effectively by accounting for uncertainty in accident likelihood and severity. In the presence of covariates, a variety of regression-type models are often used for modeling insurance claims, ranging from relatively simple generalized linear models (GLMs) to regularized GLMs to gradient boosting models (GBMs). Conformal predictive inference has arisen as a popular distribution-free approach for quantifying predictive uncertainty under relatively weak assumptions of exchangeability, and has been well studied under the classic linear regression setting. In this work, we propose new non-conformity measures for GLMs and GBMs with GLM-type loss. Using regularized Tweedie GLM regression and LightGBM with Tweedie loss, we demonstrate conformal prediction performance with these non-conformity measures in insurance claims data. Our simulation results favor the use of locally weighted Pearson residuals for LightGBM over other methods considered, as the resulting intervals maintained the nominal coverage with the smallest average width.
Interval Estimation of Coefficients in Penalized Regression Models of Insurance Data
Oct 01, 2024Abstract:The Tweedie exponential dispersion family is a popular choice among many to model insurance losses that consist of zero-inflated semicontinuous data. In such data, it is often important to obtain credibility (inference) of the most important features that describe the endogenous variables. Post-selection inference is the standard procedure in statistics to obtain confidence intervals of model parameters after performing a feature extraction procedure. For a linear model, the lasso estimate often has non-negligible estimation bias for large coefficients corresponding to exogenous variables. To have valid inference on those coefficients, it is necessary to correct the bias of the lasso estimate. Traditional statistical methods, such as hypothesis testing or standard confidence interval construction might lead to incorrect conclusions during post-selection, as they are generally too optimistic. Here we discuss a few methodologies for constructing confidence intervals of the coefficients after feature selection in the Generalized Linear Model (GLM) family with application to insurance data.
Improving Opioid Use Disorder Risk Modelling through Behavioral and Genetic Feature Integration
Sep 19, 2023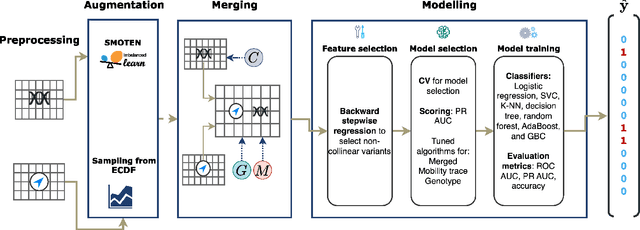
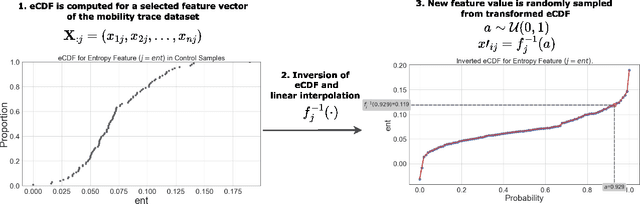
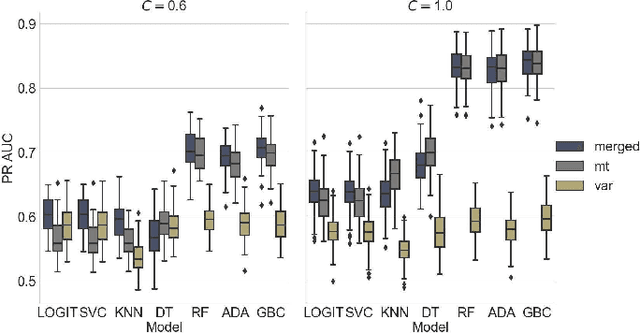
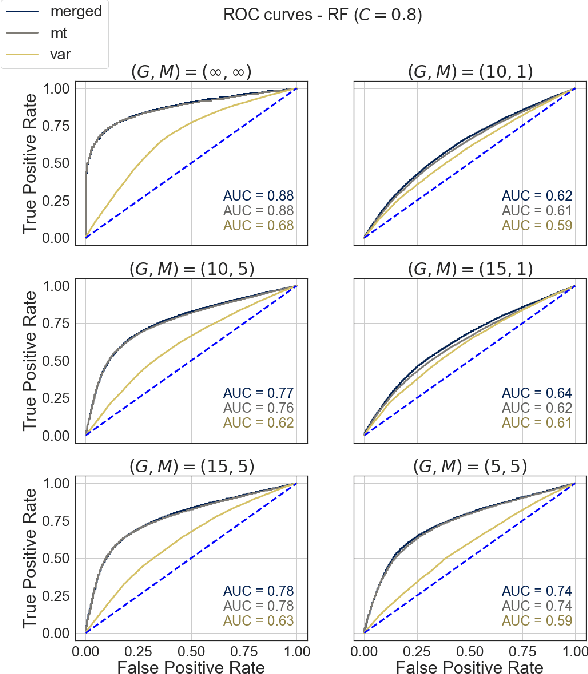
Abstract:Opioids are an effective analgesic for acute and chronic pain, but also carry a considerable risk of addiction leading to millions of opioid use disorder (OUD) cases and tens of thousands of premature deaths in the United States yearly. Estimating OUD risk prior to prescription could improve the efficacy of treatment regimens, monitoring programs, and intervention strategies, but risk estimation is typically based on self-reported data or questionnaires. We develop an experimental design and computational methods that combines genetic variants associated with OUD with behavioral features extracted from GPS and Wi-Fi spatiotemporal coordinates to assess OUD risk. Since both OUD mobility and genetic data do not exist for the same cohort, we develop algorithms to (1) generate mobility features from empirical distributions and (2) synthesize mobility and genetic samples assuming a level of comorbidity and relative risks. We show that integrating genetic and mobility modalities improves risk modelling using classification accuracy, area under the precision-recall and receiver operator characteristic curves, and $F_1$ score. Interpreting the fitted models suggests that mobility features have more influence on OUD risk, although the genetic contribution was significant, particularly in linear models. While there exists concerns with respect to privacy, security, bias, and generalizability that must be evaluated in clinical trials before being implemented in practice, our framework provides preliminary evidence that behavioral and genetic features may improve OUD risk estimation to assist with personalized clinical decision-making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge