Dimitris Pappas
Data Augmentation for Biomedical Factoid Question Answering
Apr 10, 2022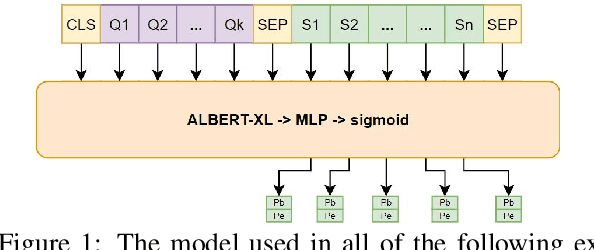

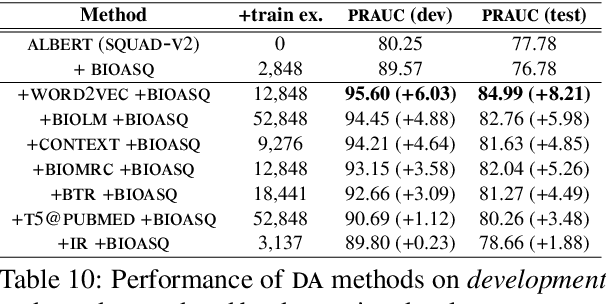

Abstract:We study the effect of seven data augmentation (da) methods in factoid question answering, focusing on the biomedical domain, where obtaining training instances is particularly difficult. We experiment with data from the BioASQ challenge, which we augment with training instances obtained from an artificial biomedical machine reading comprehension dataset, or via back-translation, information retrieval, word substitution based on word2vec embeddings, or masked language modeling, question generation, or extending the given passage with additional context. We show that da can lead to very significant performance gains, even when using large pre-trained Transformers, contributing to a broader discussion of if/when da benefits large pre-trained models. One of the simplest da methods, word2vec-based word substitution, performed best and is recommended. We release our artificial training instances and code.
A Neural Model for Joint Document and Snippet Ranking in Question Answering for Large Document Collections
Jun 16, 2021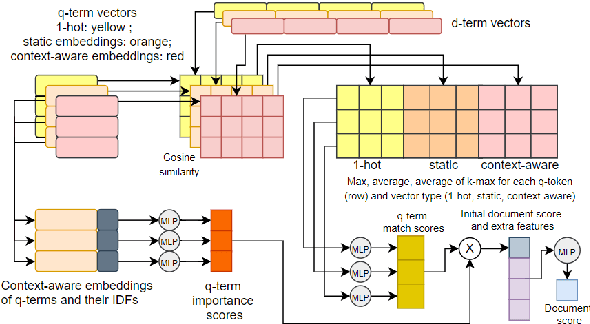

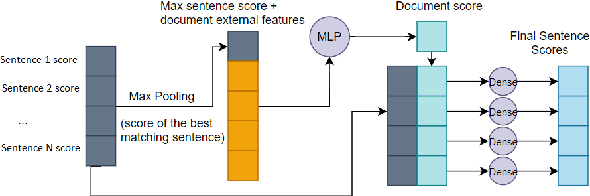

Abstract:Question answering (QA) systems for large document collections typically use pipelines that (i) retrieve possibly relevant documents, (ii) re-rank them, (iii) rank paragraphs or other snippets of the top-ranked documents, and (iv) select spans of the top-ranked snippets as exact answers. Pipelines are conceptually simple, but errors propagate from one component to the next, without later components being able to revise earlier decisions. We present an architecture for joint document and snippet ranking, the two middle stages, which leverages the intuition that relevant documents have good snippets and good snippets come from relevant documents. The architecture is general and can be used with any neural text relevance ranker. We experiment with two main instantiations of the architecture, based on POSIT-DRMM (PDRMM) and a BERT-based ranker. Experiments on biomedical data from BIOASQ show that our joint models vastly outperform the pipelines in snippet retrieval, the main goal for QA, with fewer trainable parameters, also remaining competitive in document retrieval. Furthermore, our joint PDRMM-based model is competitive with BERT-based models, despite using orders of magnitude fewer parameters. These claims are also supported by human evaluation on two test batches of BIOASQ. To test our key findings on another dataset, we modified the Natural Questions dataset so that it can also be used for document and snippet retrieval. Our joint PDRMM-based model again outperforms the corresponding pipeline in snippet retrieval on the modified Natural Questions dataset, even though it performs worse than the pipeline in document retrieval. We make our code and the modified Natural Questions dataset publicly available.
BIOMRC: A Dataset for Biomedical Machine Reading Comprehension
May 13, 2020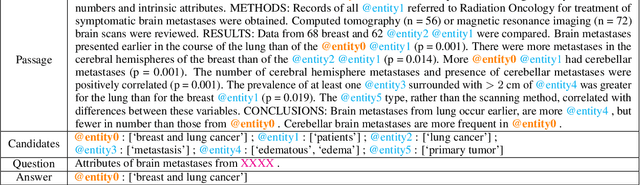
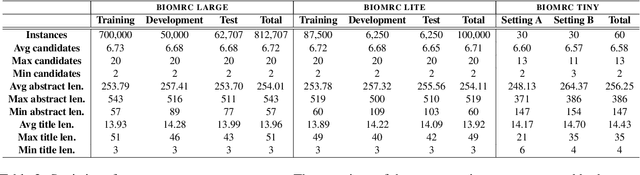
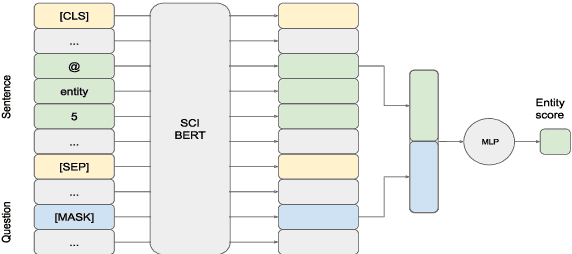
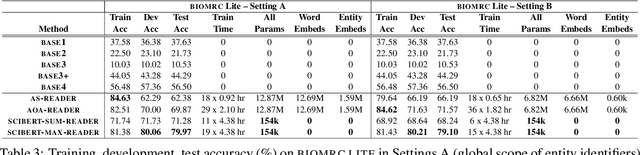
Abstract:We introduce BIOMRC, a large-scale cloze-style biomedical MRC dataset. Care was taken to reduce noise, compared to the previous BIOREAD dataset of Pappas et al. (2018). Experiments show that simple heuristics do not perform well on the new dataset, and that two neural MRC models that had been tested on BIOREAD perform much better on BIOMRC, indicating that the new dataset is indeed less noisy or at least that its task is more feasible. Non-expert human performance is also higher on the new dataset compared to BIOREAD, and biomedical experts perform even better. We also introduce a new BERT-based MRC model, the best version of which substantially outperforms all other methods tested, reaching or surpassing the accuracy of biomedical experts in some experiments. We make the new dataset available in three different sizes, also releasing our code, and providing a leaderboard.
Embedding Biomedical Ontologies by Jointly Encoding Network Structure and Textual Node Descriptors
Jun 20, 2019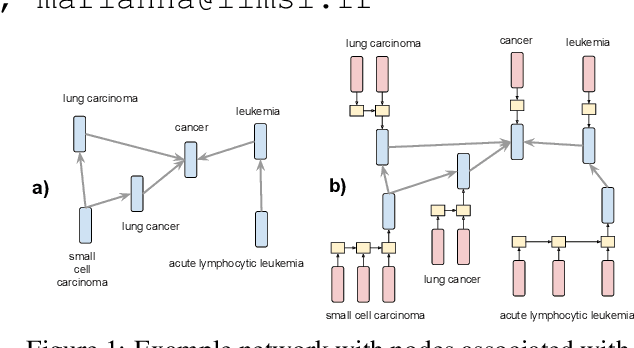
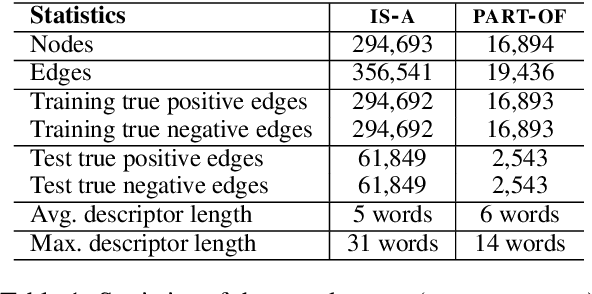
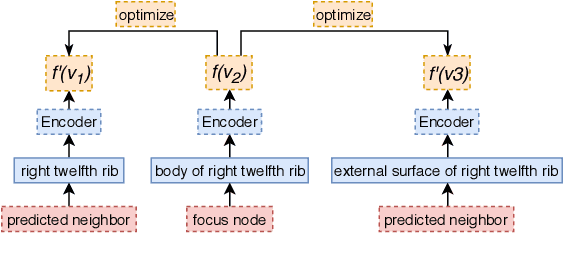
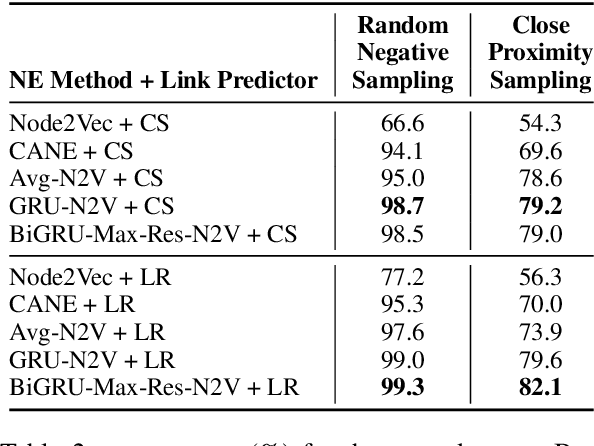
Abstract:Network Embedding (NE) methods, which map network nodes to low-dimensional feature vectors, have wide applications in network analysis and bioinformatics. Many existing NE methods rely only on network structure, overlooking other information associated with the nodes, e.g., text describing the nodes. Recent attempts to combine the two sources of information only consider local network structure. We extend NODE2VEC, a well-known NE method that considers broader network structure, to also consider textual node descriptors using recurrent neural encoders. Our method is evaluated on link prediction in two networks derived from UMLS. Experimental results demonstrate the effectiveness of the proposed approach compared to previous work.
AUEB at BioASQ 6: Document and Snippet Retrieval
Sep 15, 2018
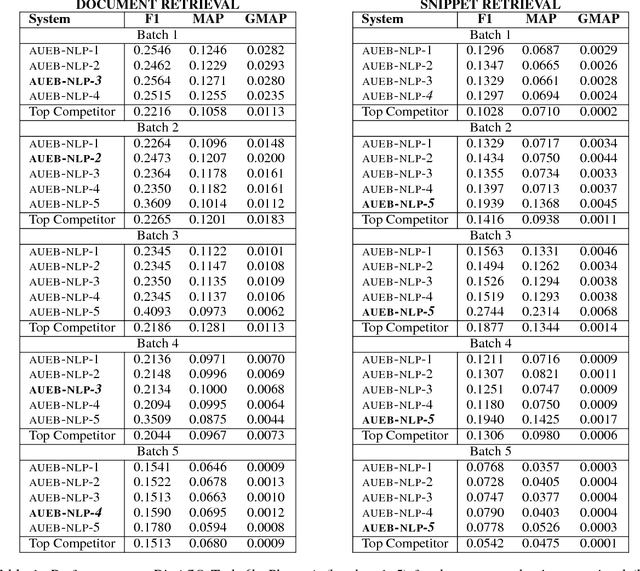

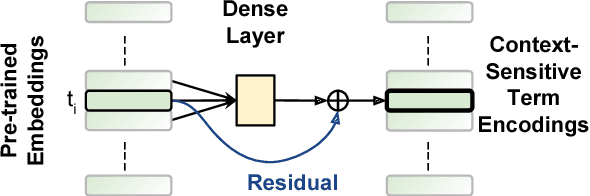
Abstract:We present AUEB's submissions to the BioASQ 6 document and snippet retrieval tasks (parts of Task 6b, Phase A). Our models use novel extensions to deep learning architectures that operate solely over the text of the query and candidate document/snippets. Our systems scored at the top or near the top for all batches of the challenge, highlighting the effectiveness of deep learning for these tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge