Dhruv V. Patel
Multi-fidelity Hamiltonian Monte Carlo
May 08, 2024Abstract:Numerous applications in biology, statistics, science, and engineering require generating samples from high-dimensional probability distributions. In recent years, the Hamiltonian Monte Carlo (HMC) method has emerged as a state-of-the-art Markov chain Monte Carlo technique, exploiting the shape of such high-dimensional target distributions to efficiently generate samples. Despite its impressive empirical success and increasing popularity, its wide-scale adoption remains limited due to the high computational cost of gradient calculation. Moreover, applying this method is impossible when the gradient of the posterior cannot be computed (for example, with black-box simulators). To overcome these challenges, we propose a novel two-stage Hamiltonian Monte Carlo algorithm with a surrogate model. In this multi-fidelity algorithm, the acceptance probability is computed in the first stage via a standard HMC proposal using an inexpensive differentiable surrogate model, and if the proposal is accepted, the posterior is evaluated in the second stage using the high-fidelity (HF) numerical solver. Splitting the standard HMC algorithm into these two stages allows for approximating the gradient of the posterior efficiently, while producing accurate posterior samples by using HF numerical solvers in the second stage. We demonstrate the effectiveness of this algorithm for a range of problems, including linear and nonlinear Bayesian inverse problems with in-silico data and experimental data. The proposed algorithm is shown to seamlessly integrate with various low-fidelity and HF models, priors, and datasets. Remarkably, our proposed method outperforms the traditional HMC algorithm in both computational and statistical efficiency by several orders of magnitude, all while retaining or improving the accuracy in computed posterior statistics.
The efficacy and generalizability of conditional GANs for posterior inference in physics-based inverse problems
Feb 15, 2022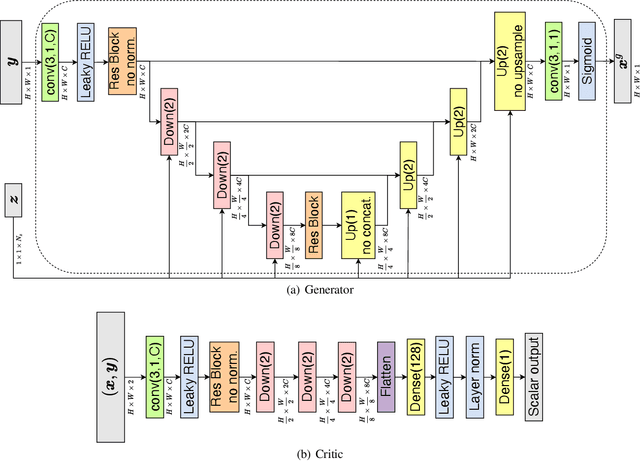
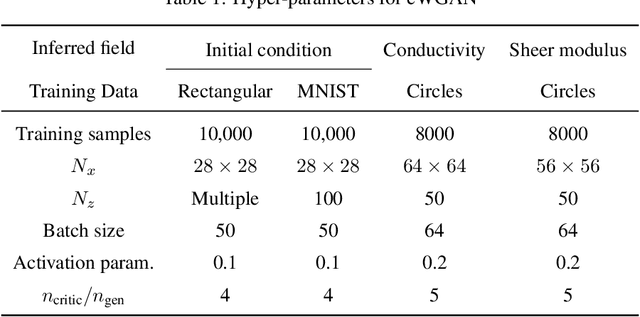
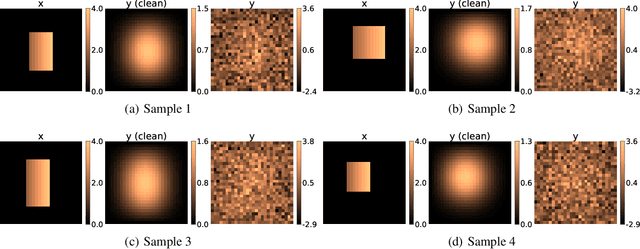
Abstract:In this work, we train conditional Wasserstein generative adversarial networks to effectively sample from the posterior of physics-based Bayesian inference problems. The generator is constructed using a U-Net architecture, with the latent information injected using conditional instance normalization. The former facilitates a multiscale inverse map, while the latter enables the decoupling of the latent space dimension from the dimension of the measurement, and introduces stochasticity at all scales of the U-Net. We solve PDE-based inverse problems to demonstrate the performance of our approach in quantifying the uncertainty in the inferred field. Further, we show the generator can learn inverse maps which are local in nature, which in turn promotes generalizability when testing with out-of-distribution samples.
GAN-based Priors for Quantifying Uncertainty
Mar 27, 2020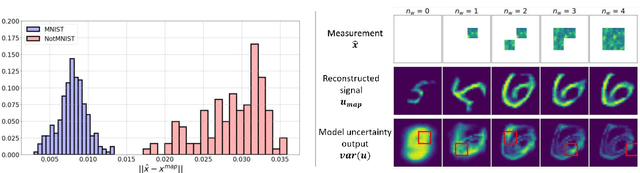
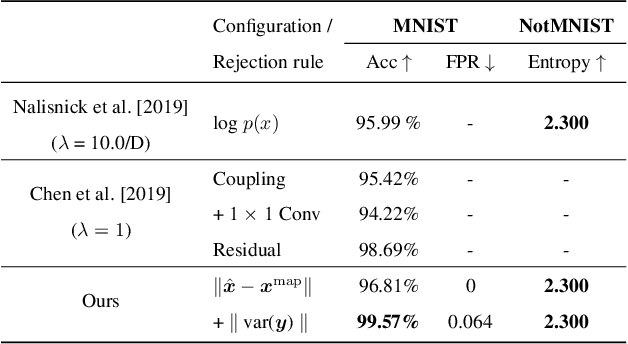
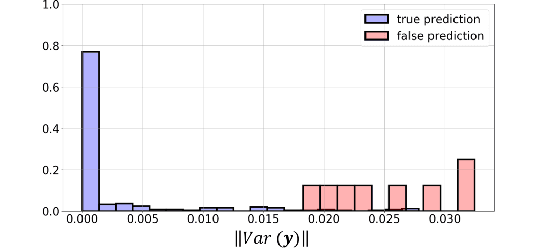
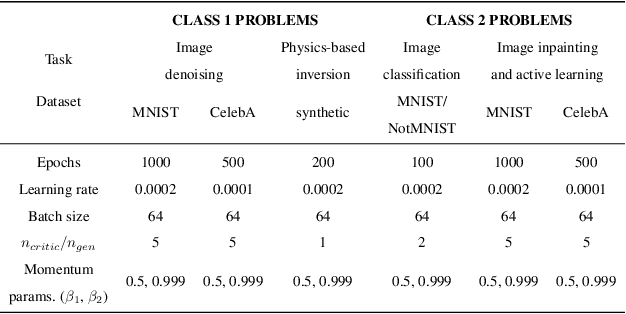
Abstract:Bayesian inference is used extensively to quantify the uncertainty in an inferred field given the measurement of a related field when the two are linked by a mathematical model. Despite its many applications, Bayesian inference faces challenges when inferring fields that have discrete representations of large dimension, and/or have prior distributions that are difficult to characterize mathematically. In this work we demonstrate how the approximate distribution learned by a deep generative adversarial network (GAN) may be used as a prior in a Bayesian update to address both these challenges. We demonstrate the efficacy of this approach on two distinct, and remarkably broad, classes of problems. The first class leads to supervised learning algorithms for image classification with superior out of distribution detection and accuracy, and for image inpainting with built-in variance estimation. The second class leads to unsupervised learning algorithms for image denoising and for solving physics-driven inverse problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge