Davide Moroni
Batch-CAM: Introduction to better reasoning in convolutional deep learning models
Oct 01, 2025Abstract:Understanding the inner workings of deep learning models is crucial for advancing artificial intelligence, particularly in high-stakes fields such as healthcare, where accurate explanations are as vital as precision. This paper introduces Batch-CAM, a novel training paradigm that fuses a batch implementation of the Grad-CAM algorithm with a prototypical reconstruction loss. This combination guides the model to focus on salient image features, thereby enhancing its performance across classification tasks. Our results demonstrate that Batch-CAM achieves a simultaneous improvement in accuracy and image reconstruction quality while reducing training and inference times. By ensuring models learn from evidence-relevant information,this approach makes a relevant contribution to building more transparent, explainable, and trustworthy AI systems.
A Classification-Aware Super-Resolution Framework for Ship Targets in SAR Imagery
Aug 08, 2025Abstract:High-resolution imagery plays a critical role in improving the performance of visual recognition tasks such as classification, detection, and segmentation. In many domains, including remote sensing and surveillance, low-resolution images can limit the accuracy of automated analysis. To address this, super-resolution (SR) techniques have been widely adopted to attempt to reconstruct high-resolution images from low-resolution inputs. Related traditional approaches focus solely on enhancing image quality based on pixel-level metrics, leaving the relationship between super-resolved image fidelity and downstream classification performance largely underexplored. This raises a key question: can integrating classification objectives directly into the super-resolution process further improve classification accuracy? In this paper, we try to respond to this question by investigating the relationship between super-resolution and classification through the deployment of a specialised algorithmic strategy. We propose a novel methodology that increases the resolution of synthetic aperture radar imagery by optimising loss functions that account for both image quality and classification performance. Our approach improves image quality, as measured by scientifically ascertained image quality indicators, while also enhancing classification accuracy.
Feature-Space Oversampling for Addressing Class Imbalance in SAR Ship Classification
Aug 08, 2025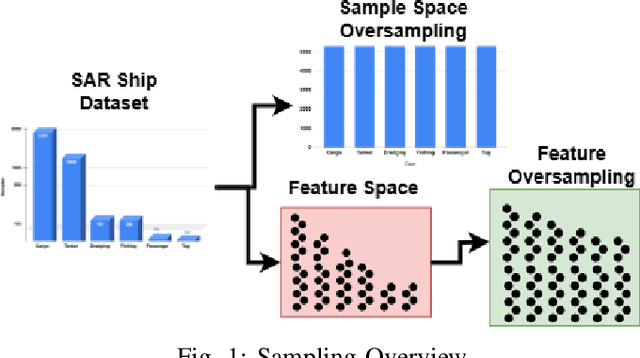
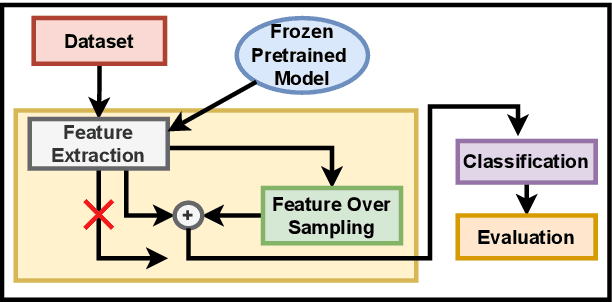
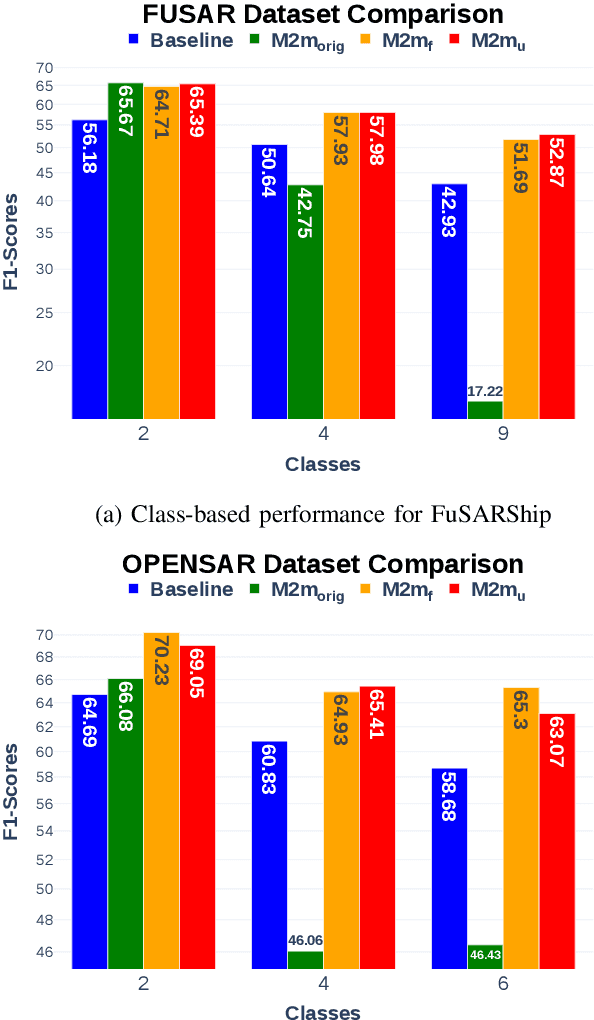
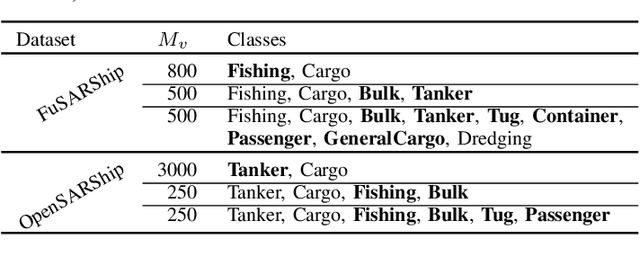
Abstract:SAR ship classification faces the challenge of long-tailed datasets, which complicates the classification of underrepresented classes. Oversampling methods have proven effective in addressing class imbalance in optical data. In this paper, we evaluated the effect of oversampling in the feature space for SAR ship classification. We propose two novel algorithms inspired by the Major-to-minor (M2m) method M2m$_f$, M2m$_u$. The algorithms are tested on two public datasets, OpenSARShip (6 classes) and FuSARShip (9 classes), using three state-of-the-art models as feature extractors: ViT, VGG16, and ResNet50. Additionally, we also analyzed the impact of oversampling methods on different class sizes. The results demonstrated the effectiveness of our novel methods over the original M2m and baselines, with an average F1-score increase of 8.82% for FuSARShip and 4.44% for OpenSARShip.
A Survey on SAR ship classification using Deep Learning
Mar 14, 2025Abstract:Deep learning (DL) has emerged as a powerful tool for Synthetic Aperture Radar (SAR) ship classification. This survey comprehensively analyzes the diverse DL techniques employed in this domain. We identify critical trends and challenges, highlighting the importance of integrating handcrafted features, utilizing public datasets, data augmentation, fine-tuning, explainability techniques, and fostering interdisciplinary collaborations to improve DL model performance. This survey establishes a first-of-its-kind taxonomy for categorizing relevant research based on DL models, handcrafted feature use, SAR attribute utilization, and the impact of fine-tuning. We discuss the methodologies used in SAR ship classification tasks and the impact of different techniques. Finally, the survey explores potential avenues for future research, including addressing data scarcity, exploring novel DL architectures, incorporating interpretability techniques, and establishing standardized performance metrics. By addressing these challenges and leveraging advancements in DL, researchers can contribute to developing more accurate and efficient ship classification systems, ultimately enhancing maritime surveillance and related applications.
A Topological Machine Learning Pipeline for Classification
Sep 26, 2023Abstract:In this work, we develop a pipeline that associates Persistence Diagrams to digital data via the most appropriate filtration for the type of data considered. Using a grid search approach, this pipeline determines optimal representation methods and parameters. The development of such a topological pipeline for Machine Learning involves two crucial steps that strongly affect its performance: firstly, digital data must be represented as an algebraic object with a proper associated filtration in order to compute its topological summary, the Persistence Diagram. Secondly, the persistence diagram must be transformed with suitable representation methods in order to be introduced in a Machine Learning algorithm. We assess the performance of our pipeline, and in parallel, we compare the different representation methods on popular benchmark datasets. This work is a first step toward both an easy and ready-to-use pipeline for data classification using persistent homology and Machine Learning, and to understand the theoretical reasons why, given a dataset and a task to be performed, a pair (filtration, topological representation) is better than another.
Alzheimer Disease Detection from Raman Spectroscopy of the Cerebrospinal Fluid via Topological Machine Learning
Sep 07, 2023



Abstract:The cerebrospinal fluid (CSF) of 19 subjects who received a clinical diagnosis of Alzheimer's disease (AD) as well as of 5 pathological controls have been collected and analysed by Raman spectroscopy (RS). We investigated whether the raw and preprocessed Raman spectra could be used to distinguish AD from controls. First, we applied standard Machine Learning (ML) methods obtaining unsatisfactory results. Then, we applied ML to a set of topological descriptors extracted from raw spectra, achieving a very good classification accuracy (>87%). Although our results are preliminary, they indicate that RS and topological analysis together may provide an effective combination to confirm or disprove a clinical diagnosis of AD. The next steps will include enlarging the dataset of CSF samples to validate the proposed method better and, possibly, to understand if topological data analysis could support the characterization of AD subtypes.
Are We Using Autoencoders in a Wrong Way?
Sep 04, 2023Abstract:Autoencoders are certainly among the most studied and used Deep Learning models: the idea behind them is to train a model in order to reconstruct the same input data. The peculiarity of these models is to compress the information through a bottleneck, creating what is called Latent Space. Autoencoders are generally used for dimensionality reduction, anomaly detection and feature extraction. These models have been extensively studied and updated, given their high simplicity and power. Examples are (i) the Denoising Autoencoder, where the model is trained to reconstruct an image from a noisy one; (ii) Sparse Autoencoder, where the bottleneck is created by a regularization term in the loss function; (iii) Variational Autoencoder, where the latent space is used to generate new consistent data. In this article, we revisited the standard training for the undercomplete Autoencoder modifying the shape of the latent space without using any explicit regularization term in the loss function. We forced the model to reconstruct not the same observation in input, but another one sampled from the same class distribution. We also explored the behaviour of the latent space in the case of reconstruction of a random sample from the whole dataset.
High Dynamic Range Imaging via Visual Attention Modules
Jul 27, 2023Abstract:Thanks to High Dynamic Range (HDR) imaging methods, the scope of photography has seen profound changes recently. To be more specific, such methods try to reconstruct the lost luminosity of the real world caused by the limitation of regular cameras from the Low Dynamic Range (LDR) images. Additionally, although the State-Of-The-Art methods in this topic perform well, they mainly concentrate on combining different exposures and have less attention to extracting the informative parts of the images. Thus, this paper aims to introduce a new model capable of incorporating information from the most visible areas of each image extracted by a visual attention module (VAM), which is a result of a segmentation strategy. In particular, the model, based on a deep learning architecture, utilizes the extracted areas to produce the final HDR image. The results demonstrate that our method outperformed most of the State-Of-The-Art algorithms.
Supervised Image Segmentation for High Dynamic Range Imaging
Dec 06, 2022Abstract:Regular cameras and cell phones are able to capture limited luminosity. Thus, in terms of quality, most of the produced images from such devices are not similar to the real world. They are overly dark or too bright, and the details are not perfectly visible. Various methods, which fall under the name of High Dynamic Range (HDR) Imaging, can be utilised to cope with this problem. Their objective is to produce an image with more details. However, unfortunately, most methods for generating an HDR image from Multi-Exposure images only concentrate on how to combine different exposures and do not have any focus on choosing the best details of each image. Therefore, it is strived in this research to extract the most visible areas of each image with the help of image segmentation. Two methods of producing the Ground Truth were considered, as manual threshold and Otsu threshold, and a neural network will be used to train segment these areas. Finally, it will be shown that the neural network is able to segment the visible parts of pictures acceptably.
Efficient Adaptive Ensembling for Image Classification
Jun 15, 2022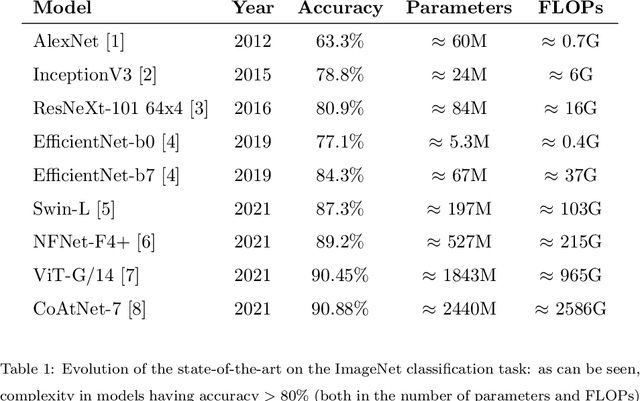
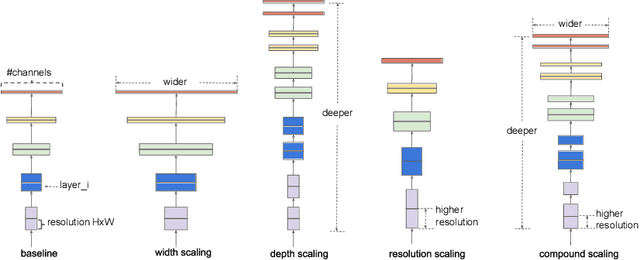
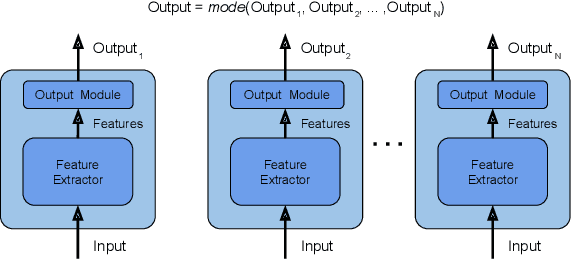
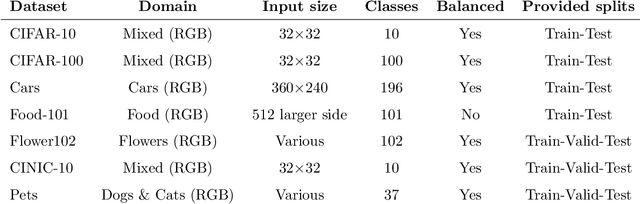
Abstract:In recent times, except for sporadic cases, the trend in Computer Vision is to achieve minor improvements over considerable increases in complexity. To reverse this tendency, we propose a novel method to boost image classification performances without an increase in complexity. To this end, we revisited ensembling, a powerful approach, not often adequately used due to its nature of increased complexity and training time, making it viable by specific design choices. First, we trained end-to-end two EfficientNet-b0 models (known to be the architecture with the best overall accuracy/complexity trade-off in image classification) on disjoint subsets of data (i.e. bagging). Then, we made an efficient adaptive ensemble by performing fine-tuning of a trainable combination layer. In this way, we were able to outperform the state-of-the-art by an average of 0.5\% on the accuracy with restrained complexity both in terms of number of parameters (by 5-60 times), and FLoating point Operations Per Second (by 10-100 times) on several major benchmark datasets, fully embracing the green AI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge