Daniele Loiacono
Memory-Efficient 3D High-Resolution Medical Image Synthesis Using CRF-Guided GANs
Mar 13, 2025Abstract:Generative Adversarial Networks (GANs) have many potential medical imaging applications. Due to the limited memory of Graphical Processing Units (GPUs), most current 3D GAN models are trained on low-resolution medical images, these models cannot scale to high-resolution or are susceptible to patchy artifacts. In this work, we propose an end-to-end novel GAN architecture that uses Conditional Random field (CRF) to model dependencies so that it can generate consistent 3D medical Images without exploiting memory. To achieve this purpose, the generator is divided into two parts during training, the first part produces an intermediate representation and CRF is applied to this intermediate representation to capture correlations. The second part of the generator produces a random sub-volume of image using a subset of the intermediate representation. This structure has two advantages: first, the correlations are modeled by using the features that the generator is trying to optimize. Second, the generator can generate full high-resolution images during inference. Experiments on Lung CTs and Brain MRIs show that our architecture outperforms state-of-the-art while it has lower memory usage and less complexity.
ARTInp: CBCT-to-CT Image Inpainting and Image Translation in Radiotherapy
Feb 07, 2025



Abstract:A key step in Adaptive Radiation Therapy (ART) workflows is the evaluation of the patient's anatomy at treatment time to ensure the accuracy of the delivery. To this end, Cone Beam Computerized Tomography (CBCT) is widely used being cost-effective and easy to integrate into the treatment process. Nonetheless, CBCT images have lower resolution and more artifacts than CT scans, making them less reliable for precise treatment validation. Moreover, in complex treatments such as Total Marrow and Lymph Node Irradiation (TMLI), where full-body visualization of the patient is critical for accurate dose delivery, the CBCT images are often discontinuous, leaving gaps that could contain relevant anatomical information. To address these limitations, we propose ARTInp (Adaptive Radiation Therapy Inpainting), a novel deep-learning framework combining image inpainting and CBCT-to-CT translation. ARTInp employs a dual-network approach: a completion network that fills anatomical gaps in CBCT volumes and a custom Generative Adversarial Network (GAN) to generate high-quality synthetic CT (sCT) images. We trained ARTInp on a dataset of paired CBCT and CT images from the SynthRad 2023 challenge, and the performance achieved on a test set of 18 patients demonstrates its potential for enhancing CBCT-based workflows in radiotherapy.
Comparative clinical evaluation of "memory-efficient" synthetic 3d generative adversarial networks (gan) head-to-head to state of art: results on computed tomography of the chest
Jan 26, 2025Abstract:Introduction: Generative Adversarial Networks (GANs) are increasingly used to generate synthetic medical images, addressing the critical shortage of annotated data for training Artificial Intelligence (AI) systems. This study introduces a novel memory-efficient GAN architecture, incorporating Conditional Random Fields (CRFs) to generate high-resolution 3D medical images and evaluates its performance against the state-of-the-art hierarchical (HA)-GAN model. Materials and Methods: The CRF-GAN was trained using the open-source lung CT LUNA16 dataset. The architecture was compared to HA-GAN through a quantitative evaluation, using Frechet Inception Distance (FID) and Maximum Mean Discrepancy (MMD) metrics, and a qualitative evaluation, through a two-alternative forced choice (2AFC) test completed by a pool of 12 resident radiologists, in order to assess the realism of the generated images. Results: CRF-GAN outperformed HA-GAN with lower FID (0.047 vs. 0.061) and MMD (0.084 vs. 0.086) scores, indicating better image fidelity. The 2AFC test showed a significant preference for images generated by CRF-Gan over those generated by HA-GAN with a p-value of 1.93e-05. Additionally, CRF-GAN demonstrated 9.34% lower memory usage at 256 resolution and achieved up to 14.6% faster training speeds, offering substantial computational savings. Discussion: CRF-GAN model successfully generates high-resolution 3D medical images with non-inferior quality to conventional models, while being more memory-efficient and faster. Computational power and time saved can be used to improve the spatial resolution and anatomical accuracy of generated images, which is still a critical factor limiting their direct clinical applicability.
Investigating Gender Bias in Lymph-node Segmentation with Anatomical Priors
Sep 24, 2024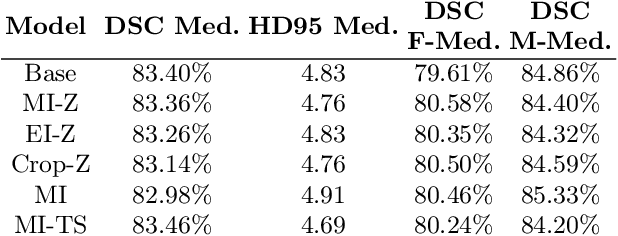
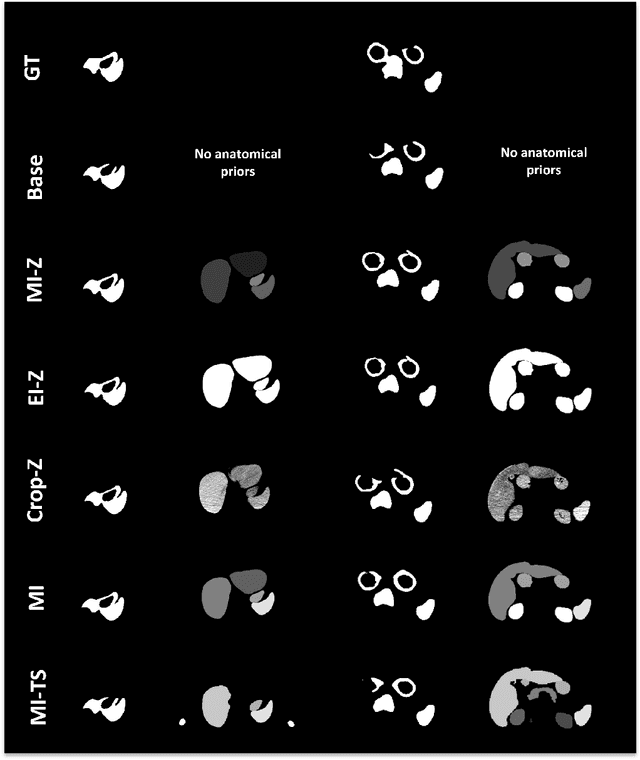
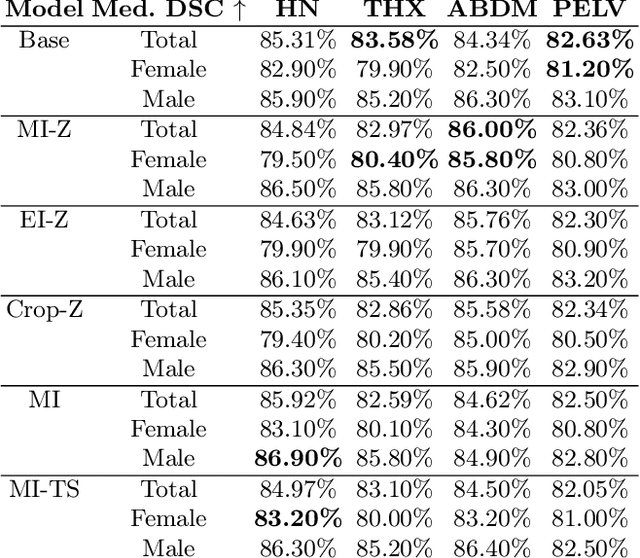
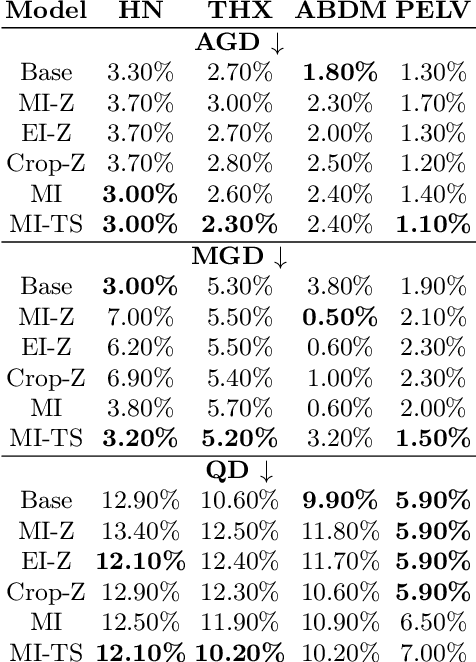
Abstract:Radiotherapy requires precise segmentation of organs at risk (OARs) and of the Clinical Target Volume (CTV) to maximize treatment efficacy and minimize toxicity. While deep learning (DL) has significantly advanced automatic contouring, complex targets like CTVs remain challenging. This study explores the use of simpler, well-segmented structures (e.g., OARs) as Anatomical Prior (AP) information to improve CTV segmentation. We investigate gender bias in segmentation models and the mitigation effect of the prior information. Findings indicate that incorporating prior knowledge with the discussed strategies enhances segmentation quality in female patients and reduces gender bias, particularly in the abdomen region. This research provides a comparative analysis of new encoding strategies and highlights the potential of using AP to achieve fairer segmentation outcomes.
Leveraging Multimodal CycleGAN for the Generation of Anatomically Accurate Synthetic CT Scans from MRIs
Jul 15, 2024



Abstract:In many clinical settings, the use of both Computed Tomography (CT) and Magnetic Resonance (MRI) is necessary to pursue a thorough understanding of the patient's anatomy and to plan a suitable therapeutical strategy; this is often the case in MRI-based radiotherapy, where CT is always necessary to prepare the dose delivery, as it provides the essential information about the radiation absorption properties of the tissues. Sometimes, MRI is preferred to contour the target volumes. However, this approach is often not the most efficient, as it is more expensive, time-consuming and, most importantly, stressful for the patients. To overcome this issue, in this work, we analyse the capabilities of different configurations of Deep Learning models to generate synthetic CT scans from MRI, leveraging the power of Generative Adversarial Networks (GANs) and, in particular, the CycleGAN architecture, capable of working in an unsupervised manner and without paired images, which were not available. Several CycleGAN models were trained unsupervised to generate CT scans from different MRI modalities with and without contrast agents. To overcome the problem of not having a ground truth, distribution-based metrics were used to assess the model's performance quantitatively, together with a qualitative evaluation where physicians were asked to differentiate between real and synthetic images to understand how realistic the generated images were. The results show how, depending on the input modalities, the models can have very different performances; however, models with the best quantitative results, according to the distribution-based metrics used, can generate very difficult images to distinguish from the real ones, even for physicians, demonstrating the approach's potential.
Deep Learning-Based Auto-Segmentation of Planning Target Volume for Total Marrow and Lymph Node Irradiation
Feb 09, 2024



Abstract:In order to optimize the radiotherapy delivery for cancer treatment, especially when dealing with complex treatments such as Total Marrow and Lymph Node Irradiation (TMLI), the accurate contouring of the Planning Target Volume (PTV) is crucial. Unfortunately, relying on manual contouring for such treatments is time-consuming and prone to errors. In this paper, we investigate the application of Deep Learning (DL) to automate the segmentation of the PTV in TMLI treatment, building upon previous work that introduced a solution to this problem based on a 2D U-Net model. We extend the previous research (i) by employing the nnU-Net framework to develop both 2D and 3D U-Net models and (ii) by evaluating the trained models on the PTV with the exclusion of bones, which consist mainly of lymp-nodes and represent the most challenging region of the target volume to segment. Our result show that the introduction of nnU-NET framework led to statistically significant improvement in the segmentation performance. In addition, the analysis on the PTV after the exclusion of bones showed that the models are quite robust also on the most challenging areas of the target volume. Overall, our study is a significant step forward in the application of DL in a complex radiotherapy treatment such as TMLI, offering a viable and scalable solution to increase the number of patients who can benefit from this treatment.
A Tool for the Procedural Generation of Shaders using Interactive Evolutionary Algorithms
Dec 29, 2023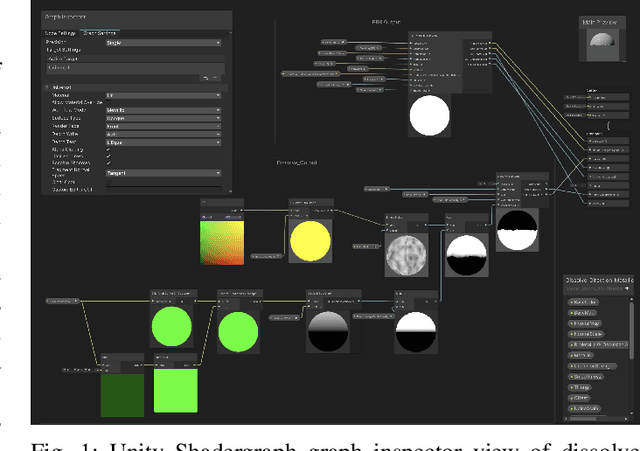
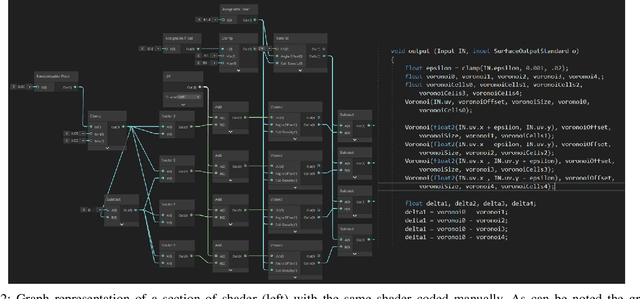
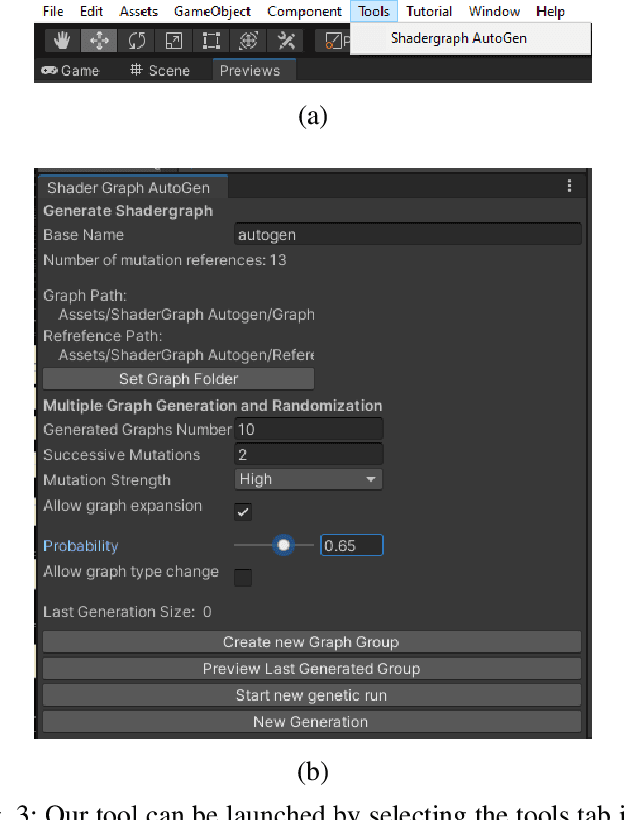
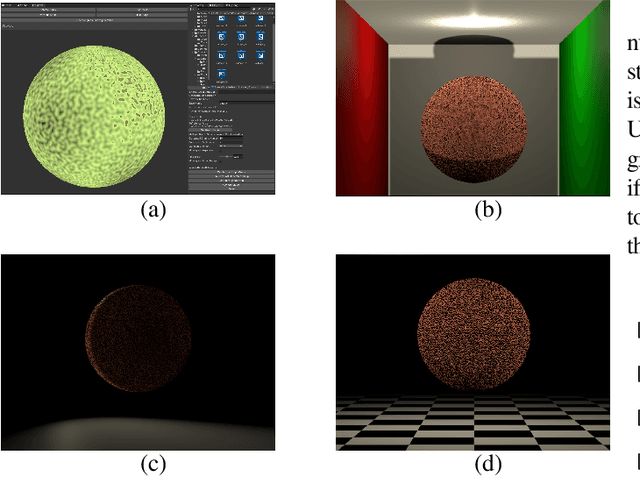
Abstract:We present a tool for exploring the design space of shaders using an interactive evolutionary algorithm integrated with the Unity editor, a well-known commercial tool for video game development. Our framework leverages the underlying graph-based representation of recent shader editors and interactive evolution to allow designers to explore several visual options starting from an existing shader. Our framework encodes the graph representation of a current shader as a chromosome used to seed the evolution of a shader population. It applies graph-based recombination and mutation with a set of heuristics to create feasible shaders. The framework is an extension of the Unity editor; thus, designers with little knowledge of evolutionary computation (and shader programming) can interact with the underlying evolutionary engine using the same visual interface used for working on game scenes.
Segmentation of Planning Target Volume in CT Series for Total Marrow Irradiation Using U-Net
Apr 05, 2023



Abstract:Radiotherapy (RT) is a key component in the treatment of various cancers, including Acute Lymphocytic Leukemia (ALL) and Acute Myelogenous Leukemia (AML). Precise delineation of organs at risk (OARs) and target areas is essential for effective treatment planning. Intensity Modulated Radiotherapy (IMRT) techniques, such as Total Marrow Irradiation (TMI) and Total Marrow and Lymph node Irradiation (TMLI), provide more precise radiation delivery compared to Total Body Irradiation (TBI). However, these techniques require time-consuming manual segmentation of structures in Computerized Tomography (CT) scans by the Radiation Oncologist (RO). In this paper, we present a deep learning-based auto-contouring method for segmenting Planning Target Volume (PTV) for TMLI treatment using the U-Net architecture. We trained and compared two segmentation models with two different loss functions on a dataset of 100 patients treated with TMLI at the Humanitas Research Hospital between 2011 and 2021. Despite challenges in lymph node areas, the best model achieved an average Dice score of 0.816 for PTV segmentation. Our findings are a preliminary but significant step towards developing a segmentation model that has the potential to save radiation oncologists a considerable amount of time. This could allow for the treatment of more patients, resulting in improved clinical practice efficiency and more reproducible contours.
Ensemble Methods for Multi-Organ Segmentation in CT Series
Mar 31, 2023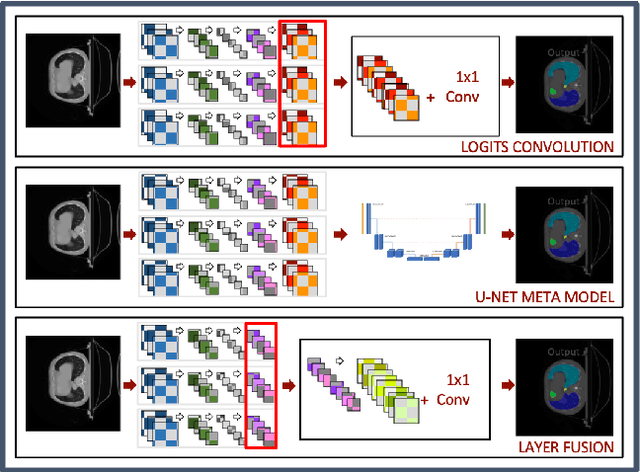
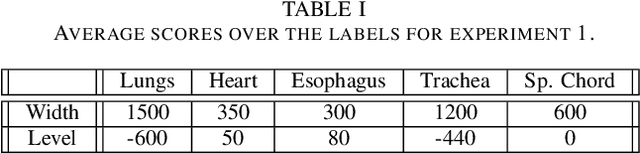
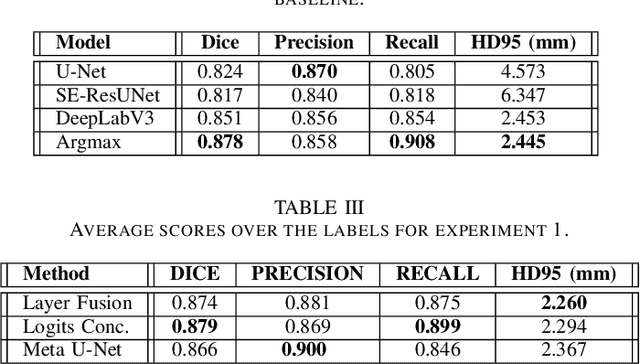
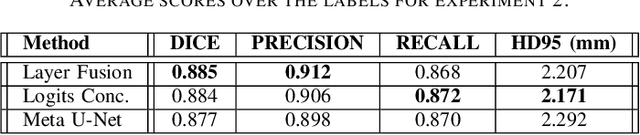
Abstract:In the medical images field, semantic segmentation is one of the most important, yet difficult and time-consuming tasks to be performed by physicians. Thanks to the recent advancement in the Deep Learning models regarding Computer Vision, the promise to automate this kind of task is getting more and more realistic. However, many problems are still to be solved, like the scarce availability of data and the difficulty to extend the efficiency of highly specialised models to general scenarios. Organs at risk segmentation for radiotherapy treatment planning falls in this category, as the limited data available negatively affects the possibility to develop general-purpose models; in this work, we focus on the possibility to solve this problem by presenting three types of ensembles of single-organ models able to produce multi-organ masks exploiting the different specialisations of their components. The results obtained are promising and prove that this is a possible solution to finding efficient multi-organ segmentation methods.
Comparing Adversarial and Supervised Learning for Organs at Risk Segmentation in CT images
Mar 31, 2023



Abstract:Organ at Risk (OAR) segmentation from CT scans is a key component of the radiotherapy treatment workflow. In recent years, deep learning techniques have shown remarkable potential in automating this process. In this paper, we investigate the performance of Generative Adversarial Networks (GANs) compared to supervised learning approaches for segmenting OARs from CT images. We propose three GAN-based models with identical generator architectures but different discriminator networks. These models are compared with well-established CNN models, such as SE-ResUnet and DeepLabV3, using the StructSeg dataset, which consists of 50 annotated CT scans containing contours of six OARs. Our work aims to provide insight into the advantages and disadvantages of adversarial training in the context of OAR segmentation. The results are very promising and show that the proposed GAN-based approaches are similar or superior to their CNN-based counterparts, particularly when segmenting more challenging target organs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge