Daniela Galimberti
The SERENADE project: Sensor-Based Explainable Detection of Cognitive Decline
Apr 11, 2025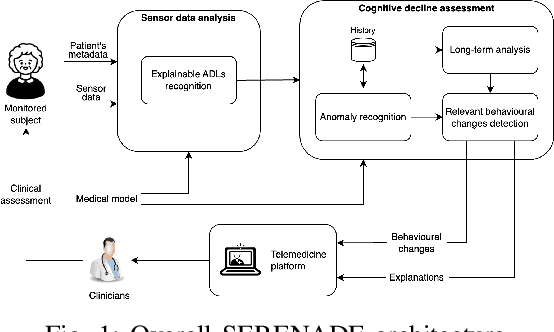
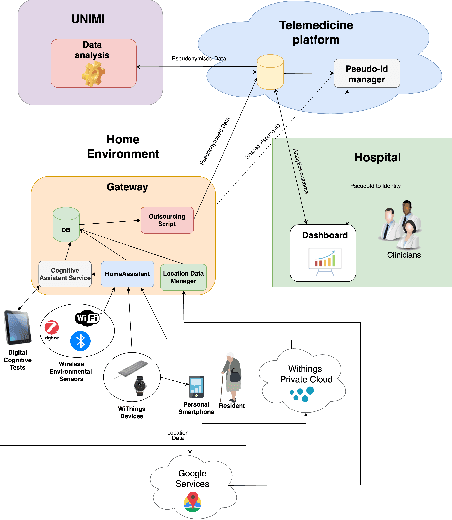
Abstract:Mild Cognitive Impairment (MCI) affects 12-18% of individuals over 60. MCI patients exhibit cognitive dysfunctions without significant daily functional loss. While MCI may progress to dementia, predicting this transition remains a clinical challenge due to limited and unreliable indicators. Behavioral changes, like in the execution of Activities of Daily Living (ADLs), can signal such progression. Sensorized smart homes and wearable devices offer an innovative solution for continuous, non-intrusive monitoring ADLs for MCI patients. However, current machine learning models for detecting behavioral changes lack transparency, hindering clinicians' trust. This paper introduces the SERENADE project, a European Union-funded initiative that aims to detect and explain behavioral changes associated with cognitive decline using explainable AI methods. SERENADE aims at collecting one year of data from 30 MCI patients living alone, leveraging AI to support clinical decision-making and offering a new approach to early dementia detection.
Identifying latent disease factors differently expressed in patient subgroups using group factor analysis
Oct 10, 2024



Abstract:In this study, we propose a novel approach to uncover subgroup-specific and subgroup-common latent factors addressing the challenges posed by the heterogeneity of neurological and mental disorders, which hinder disease understanding, treatment development, and outcome prediction. The proposed approach, sparse Group Factor Analysis (GFA) with regularised horseshoe priors, was implemented with probabilistic programming and can uncover associations (or latent factors) among multiple data modalities differentially expressed in sample subgroups. Synthetic data experiments showed the robustness of our sparse GFA by correctly inferring latent factors and model parameters. When applied to the Genetic Frontotemporal Dementia Initiative (GENFI) dataset, which comprises patients with frontotemporal dementia (FTD) with genetically defined subgroups, the sparse GFA identified latent disease factors differentially expressed across the subgroups, distinguishing between "subgroup-specific" latent factors within homogeneous groups and "subgroup common" latent factors shared across subgroups. The latent disease factors captured associations between brain structure and non-imaging variables (i.e., questionnaires assessing behaviour and disease severity) across the different genetic subgroups, offering insights into disease profiles. Importantly, two latent factors were more pronounced in the two more homogeneous FTD patient subgroups (progranulin (GRN) and microtubule-associated protein tau (MAPT) mutation), showcasing the method's ability to reveal subgroup-specific characteristics. These findings underscore the potential of sparse GFA for integrating multiple data modalities and identifying interpretable latent disease factors that can improve the characterization and stratification of patients with neurological and mental health disorders.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge