Damir Juric
Human Evaluation and Correlation with Automatic Metrics in Consultation Note Generation
Apr 01, 2022



Abstract:In recent years, machine learning models have rapidly become better at generating clinical consultation notes; yet, there is little work on how to properly evaluate the generated consultation notes to understand the impact they may have on both the clinician using them and the patient's clinical safety. To address this we present an extensive human evaluation study of consultation notes where 5 clinicians (i) listen to 57 mock consultations, (ii) write their own notes, (iii) post-edit a number of automatically generated notes, and (iv) extract all the errors, both quantitative and qualitative. We then carry out a correlation study with 18 automatic quality metrics and the human judgements. We find that a simple, character-based Levenshtein distance metric performs on par if not better than common model-based metrics like BertScore. All our findings and annotations are open-sourced.
Towards more patient friendly clinical notes through language models and ontologies
Dec 23, 2021



Abstract:Clinical notes are an efficient way to record patient information but are notoriously hard to decipher for non-experts. Automatically simplifying medical text can empower patients with valuable information about their health, while saving clinicians time. We present a novel approach to automated simplification of medical text based on word frequencies and language modelling, grounded on medical ontologies enriched with layman terms. We release a new dataset of pairs of publicly available medical sentences and a version of them simplified by clinicians. Also, we define a novel text simplification metric and evaluation framework, which we use to conduct a large-scale human evaluation of our method against the state of the art. Our method based on a language model trained on medical forum data generates simpler sentences while preserving both grammar and the original meaning, surpassing the current state of the art.
Towards objectively evaluating the quality of generated medical summaries
Apr 09, 2021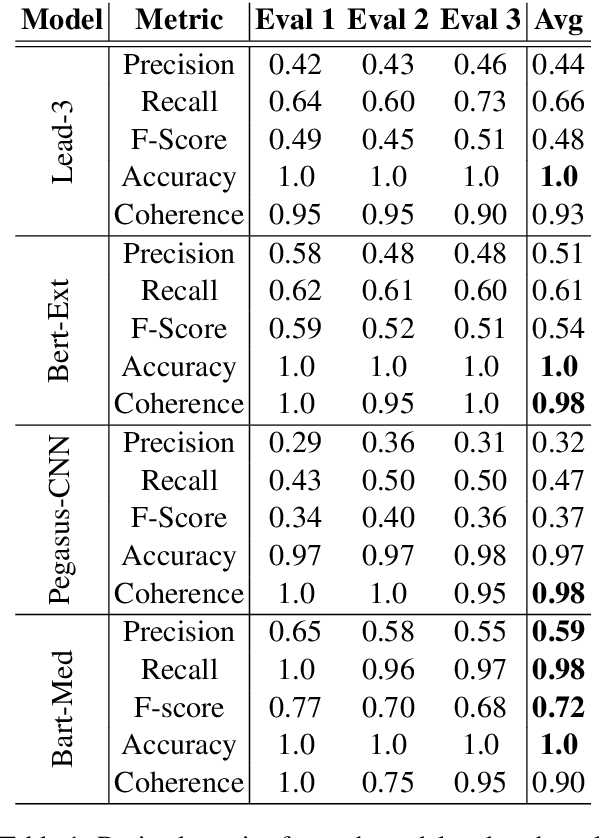

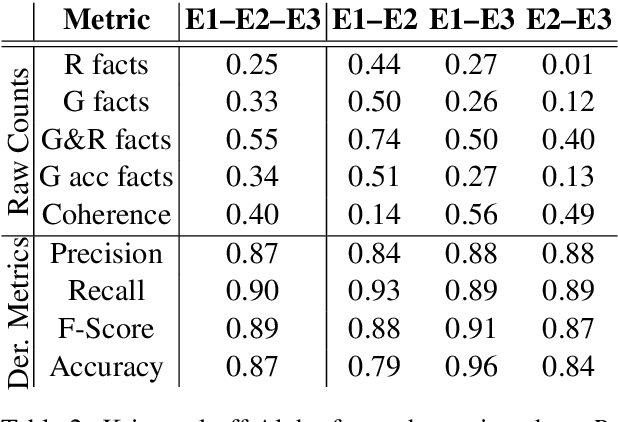

Abstract:We propose a method for evaluating the quality of generated text by asking evaluators to count facts, and computing precision, recall, f-score, and accuracy from the raw counts. We believe this approach leads to a more objective and easier to reproduce evaluation. We apply this to the task of medical report summarisation, where measuring objective quality and accuracy is of paramount importance.
Can Embeddings Adequately Represent Medical Terminology? New Large-Scale Medical Term Similarity Datasets Have the Answer!
Mar 24, 2020
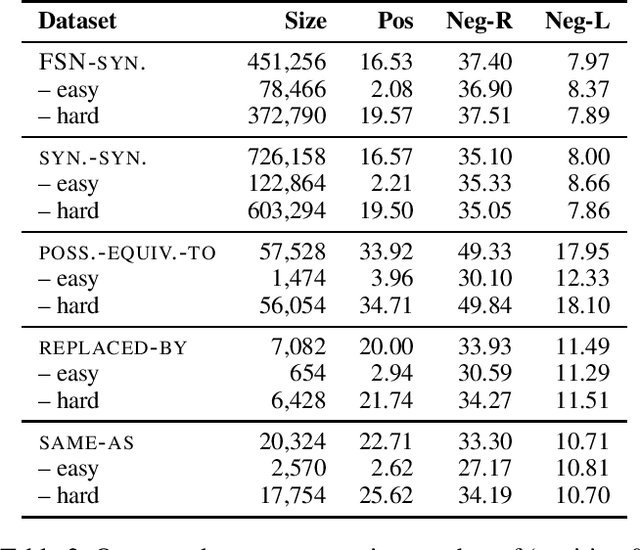
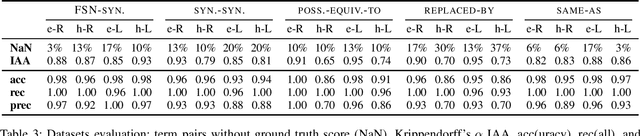
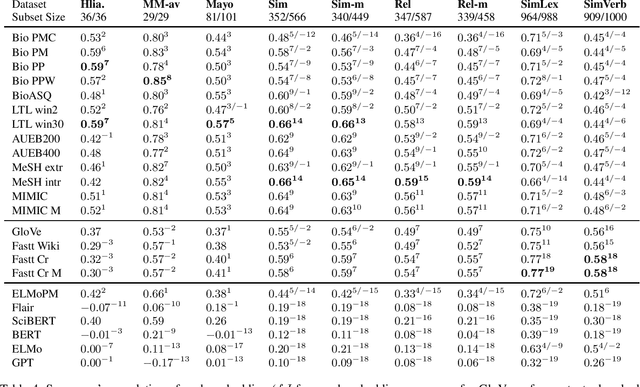
Abstract:A large number of embeddings trained on medical data have emerged, but it remains unclear how well they represent medical terminology, in particular whether the close relationship of semantically similar medical terms is encoded in these embeddings. To date, only small datasets for testing medical term similarity are available, not allowing to draw conclusions about the generalisability of embeddings to the enormous amount of medical terms used by doctors. We present multiple automatically created large-scale medical term similarity datasets and confirm their high quality in an annotation study with doctors. We evaluate state-of-the-art word and contextual embeddings on our new datasets, comparing multiple vector similarity metrics and word vector aggregation techniques. Our results show that current embeddings are limited in their ability to adequately encode medical terms. The novel datasets thus form a challenging new benchmark for the development of medical embeddings able to accurately represent the whole medical terminology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge