Connor Heaton
PEaCE: A Chemistry-Oriented Dataset for Optical Character Recognition on Scientific Documents
Mar 23, 2024Abstract:Optical Character Recognition (OCR) is an established task with the objective of identifying the text present in an image. While many off-the-shelf OCR models exist, they are often trained for either scientific (e.g., formulae) or generic printed English text. Extracting text from chemistry publications requires an OCR model that is capable in both realms. Nougat, a recent tool, exhibits strong ability to parse academic documents, but is unable to parse tables in PubMed articles, which comprises a significant part of the academic community and is the focus of this work. To mitigate this gap, we present the Printed English and Chemical Equations (PEaCE) dataset, containing both synthetic and real-world records, and evaluate the efficacy of transformer-based OCR models when trained on this resource. Given that real-world records contain artifacts not present in synthetic records, we propose transformations that mimic such qualities. We perform a suite of experiments to explore the impact of patch size, multi-domain training, and our proposed transformations, ultimately finding that models with a small patch size trained on multiple domains using the proposed transformations yield the best performance. Our dataset and code is available at https://github.com/ZN1010/PEaCE.
Leveraging External Knowledge Resources to Enable Domain-Specific Comprehension
Jan 15, 2024



Abstract:Machine Reading Comprehension (MRC) has been a long-standing problem in NLP and, with the recent introduction of the BERT family of transformer based language models, it has come a long way to getting solved. Unfortunately, however, when BERT variants trained on general text corpora are applied to domain-specific text, their performance inevitably degrades on account of the domain shift i.e. genre/subject matter discrepancy between the training and downstream application data. Knowledge graphs act as reservoirs for either open or closed domain information and prior studies have shown that they can be used to improve the performance of general-purpose transformers in domain-specific applications. Building on existing work, we introduce a method using Multi-Layer Perceptrons (MLPs) for aligning and integrating embeddings extracted from knowledge graphs with the embeddings spaces of pre-trained language models (LMs). We fuse the aligned embeddings with open-domain LMs BERT and RoBERTa, and fine-tune them for two MRC tasks namely span detection (COVID-QA) and multiple-choice questions (PubMedQA). On the COVID-QA dataset, we see that our approach allows these models to perform similar to their domain-specific counterparts, Bio/Sci-BERT, as evidenced by the Exact Match (EM) metric. With regards to PubMedQA, we observe an overall improvement in accuracy while the F1 stays relatively the same over the domain-specific models.
Quality > Quantity: Synthetic Corpora from Foundation Models for Closed-Domain Extractive Question Answering
Oct 25, 2023



Abstract:Domain adaptation, the process of training a model in one domain and applying it to another, has been extensively explored in machine learning. While training a domain-specific foundation model (FM) from scratch is an option, recent methods have focused on adapting pre-trained FMs for domain-specific tasks. However, our experiments reveal that either approach does not consistently achieve state-of-the-art (SOTA) results in the target domain. In this work, we study extractive question answering within closed domains and introduce the concept of targeted pre-training. This involves determining and generating relevant data to further pre-train our models, as opposed to the conventional philosophy of utilizing domain-specific FMs trained on a wide range of data. Our proposed framework uses Galactica to generate synthetic, ``targeted'' corpora that align with specific writing styles and topics, such as research papers and radiology reports. This process can be viewed as a form of knowledge distillation. We apply our method to two biomedical extractive question answering datasets, COVID-QA and RadQA, achieving a new benchmark on the former and demonstrating overall improvements on the latter. Code available at https://github.com/saptarshi059/CDQA-v1-Targetted-PreTraining/tree/main.
Learning To Describe Player Form in The MLB
Sep 11, 2021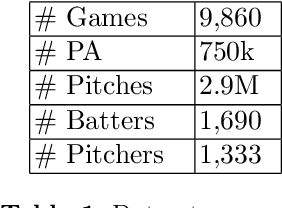
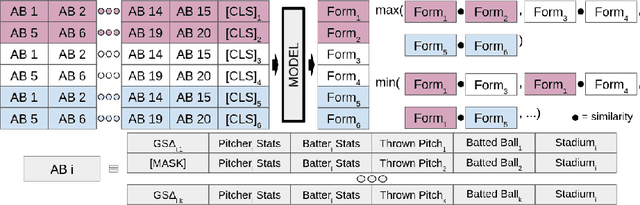
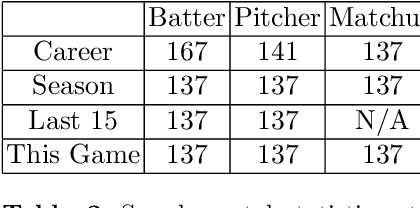
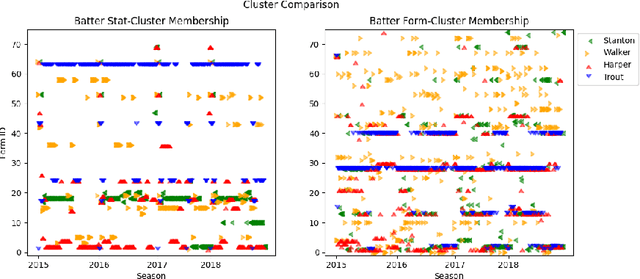
Abstract:Major League Baseball (MLB) has a storied history of using statistics to better understand and discuss the game of baseball, with an entire discipline of statistics dedicated to the craft, known as sabermetrics. At their core, all sabermetrics seek to quantify some aspect of the game, often a specific aspect of a player's skill set - such as a batter's ability to drive in runs (RBI) or a pitcher's ability to keep batters from reaching base (WHIP). While useful, such statistics are fundamentally limited by the fact that they are derived from an account of what happened on the field, not how it happened. As a first step towards alleviating this shortcoming, we present a novel, contrastive learning-based framework for describing player form in the MLB. We use form to refer to the way in which a player has impacted the course of play in their recent appearances. Concretely, a player's form is described by a 72-dimensional vector. By comparing clusters of players resulting from our form representations and those resulting from traditional abermetrics, we demonstrate that our form representations contain information about how players impact the course of play, not present in traditional, publicly available statistics. We believe these embeddings could be utilized to predict both in-game and game-level events, such as the result of an at-bat or the winner of a game.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge