Christiana Kartsonaki
At-Admission Prediction of Mortality and Pulmonary Embolism in COVID-19 Patients Using Statistical and Machine Learning Methods: An International Cohort Study
May 18, 2023Abstract:By September, 2022, more than 600 million cases of SARS-CoV-2 infection have been reported globally, resulting in over 6.5 million deaths. COVID-19 mortality risk estimators are often, however, developed with small unrepresentative samples and with methodological limitations. It is highly important to develop predictive tools for pulmonary embolism (PE) in COVID-19 patients as one of the most severe preventable complications of COVID-19. Using a dataset of more than 800,000 COVID-19 patients from an international cohort, we propose a cost-sensitive gradient-boosted machine learning model that predicts occurrence of PE and death at admission. Logistic regression, Cox proportional hazards models, and Shapley values were used to identify key predictors for PE and death. Our prediction model had a test AUROC of 75.9% and 74.2%, and sensitivities of 67.5% and 72.7% for PE and all-cause mortality respectively on a highly diverse and held-out test set. The PE prediction model was also evaluated on patients in UK and Spain separately with test results of 74.5% AUROC, 63.5% sensitivity and 78.9% AUROC, 95.7% sensitivity. Age, sex, region of admission, comorbidities (chronic cardiac and pulmonary disease, dementia, diabetes, hypertension, cancer, obesity, smoking), and symptoms (any, confusion, chest pain, fatigue, headache, fever, muscle or joint pain, shortness of breath) were the most important clinical predictors at admission. Our machine learning model developed from an international cohort can serve to better regulate hospital risk prioritisation of at-risk patients.
Using Bottleneck Adapters to Identify Cancer in Clinical Notes under Low-Resource Constraints
Oct 17, 2022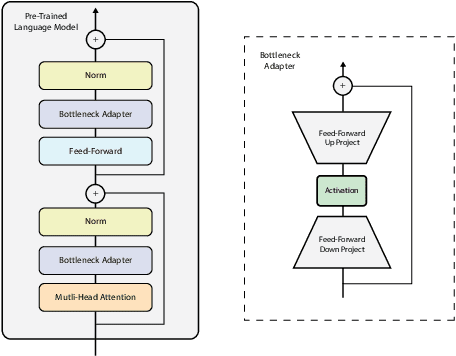
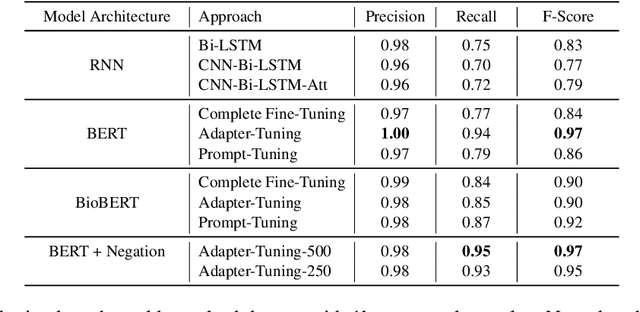
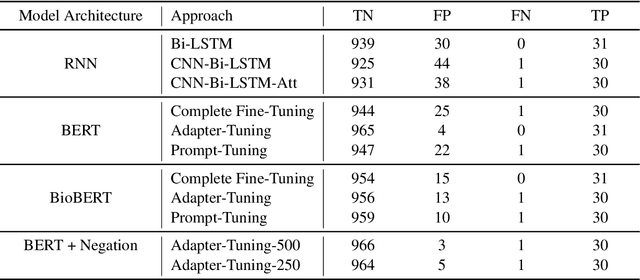
Abstract:Processing information locked within clinical health records is a challenging task that remains an active area of research in biomedical NLP. In this work, we evaluate a broad set of machine learning techniques ranging from simple RNNs to specialised transformers such as BioBERT on a dataset containing clinical notes along with a set of annotations indicating whether a sample is cancer-related or not. Furthermore, we specifically employ efficient fine-tuning methods from NLP, namely, bottleneck adapters and prompt tuning, to adapt the models to our specialised task. Our evaluations suggest that fine-tuning a frozen BERT model pre-trained on natural language and with bottleneck adapters outperforms all other strategies, including full fine-tuning of the specialised BioBERT model. Based on our findings, we suggest that using bottleneck adapters in low-resource situations with limited access to labelled data or processing capacity could be a viable strategy in biomedical text mining. The code used in the experiments are going to be made available at https://github.com/omidrohanian/bottleneck-adapters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge