Chenxi Jiang
REI-Bench: Can Embodied Agents Understand Vague Human Instructions in Task Planning?
May 19, 2025Abstract:Robot task planning decomposes human instructions into executable action sequences that enable robots to complete a series of complex tasks. Although recent large language model (LLM)-based task planners achieve amazing performance, they assume that human instructions are clear and straightforward. However, real-world users are not experts, and their instructions to robots often contain significant vagueness. Linguists suggest that such vagueness frequently arises from referring expressions (REs), whose meanings depend heavily on dialogue context and environment. This vagueness is even more prevalent among the elderly and children, who robots should serve more. This paper studies how such vagueness in REs within human instructions affects LLM-based robot task planning and how to overcome this issue. To this end, we propose the first robot task planning benchmark with vague REs (REI-Bench), where we discover that the vagueness of REs can severely degrade robot planning performance, leading to success rate drops of up to 77.9%. We also observe that most failure cases stem from missing objects in planners. To mitigate the REs issue, we propose a simple yet effective approach: task-oriented context cognition, which generates clear instructions for robots, achieving state-of-the-art performance compared to aware prompt and chains of thought. This work contributes to the research community of human-robot interaction (HRI) by making robot task planning more practical, particularly for non-expert users, e.g., the elderly and children.
UniRiT: Towards Few-Shot Non-Rigid Point Cloud Registration
Oct 30, 2024Abstract:Non-rigid point cloud registration is a critical challenge in 3D scene understanding, particularly in surgical navigation. Although existing methods achieve excellent performance when trained on large-scale, high-quality datasets, these datasets are prohibitively expensive to collect and annotate, e.g., organ data in authentic medical scenarios. With insufficient training samples and data noise, existing methods degrade significantly since non-rigid patterns are more flexible and complicated than rigid ones, and the distributions across samples are more distinct, leading to higher difficulty in representation learning with few data. In this work, we aim to deal with this challenging few-shot non-rigid point cloud registration problem. Based on the observation that complex non-rigid transformation patterns can be decomposed into rigid and small non-rigid transformations, we propose a novel and effective framework, UniRiT. UniRiT adopts a two-step registration strategy that first aligns the centroids of the source and target point clouds and then refines the registration with non-rigid transformations, thereby significantly reducing the problem complexity. To validate the performance of UniRiT on real-world datasets, we introduce a new dataset, MedMatch3D, which consists of real human organs and exhibits high variability in sample distribution. We further establish a new challenging benchmark for few-shot non-rigid registration. Extensive empirical results demonstrate that UniRiT achieves state-of-the-art performance on MedMatch3D, improving the existing best approach by 94.22%.
Enhancing Dataset Distillation via Label Inconsistency Elimination and Learning Pattern Refinement
Oct 17, 2024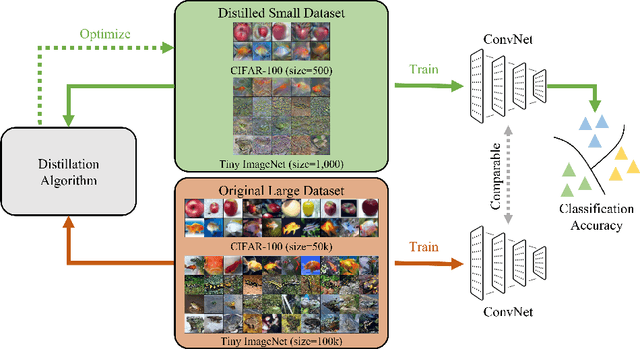
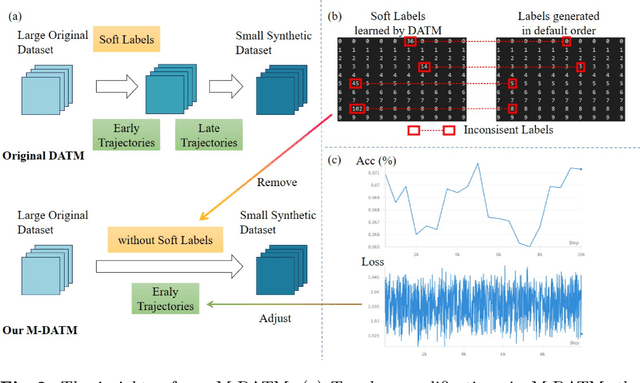
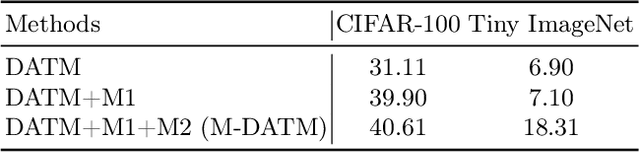
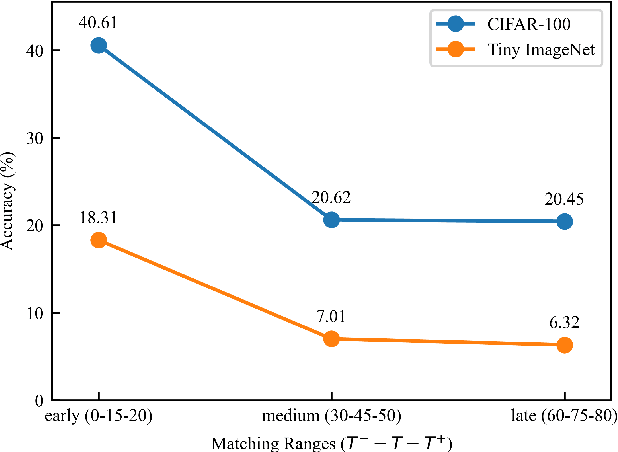
Abstract:Dataset Distillation (DD) seeks to create a condensed dataset that, when used to train a model, enables the model to achieve performance similar to that of a model trained on the entire original dataset. It relieves the model training from processing massive data and thus reduces the computation resources, storage, and time costs. This paper illustrates our solution that ranks 1st in the ECCV-2024 Data Distillation Challenge (track 1). Our solution, Modified Difficulty-Aligned Trajectory Matching (M-DATM), introduces two key modifications to the original state-of-the-art method DATM: (1) the soft labels learned by DATM do not achieve one-to-one correspondence with the counterparts generated by the official evaluation script, so we remove the soft labels technique to alleviate such inconsistency; (2) since the removal of soft labels makes it harder for the synthetic dataset to learn late trajectory information, particularly on Tiny ImageNet, we reduce the matching range, allowing the synthetic data to concentrate more on the easier patterns. In the final evaluation, our M-DATM achieved accuracies of 0.4061 and 0.1831 on the CIFAR-100 and Tiny ImageNet datasets, ranking 1st in the Fixed Images Per Class (IPC) Track.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge