Chenghui Zhou
Chemistry-Inspired Diffusion with Non-Differentiable Guidance
Oct 09, 2024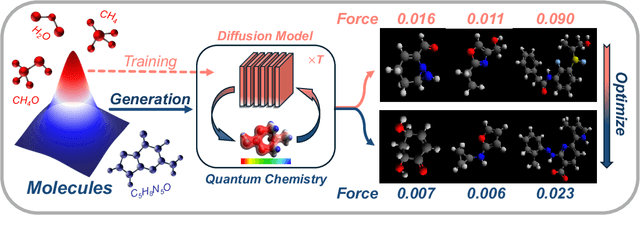

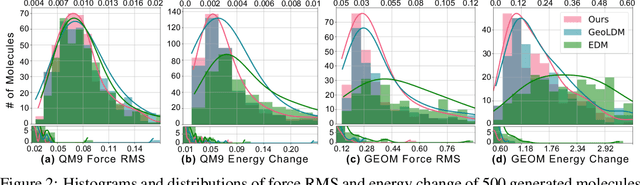

Abstract:Recent advances in diffusion models have shown remarkable potential in the conditional generation of novel molecules. These models can be guided in two ways: (i) explicitly, through additional features representing the condition, or (ii) implicitly, using a property predictor. However, training property predictors or conditional diffusion models requires an abundance of labeled data and is inherently challenging in real-world applications. We propose a novel approach that attenuates the limitations of acquiring large labeled datasets by leveraging domain knowledge from quantum chemistry as a non-differentiable oracle to guide an unconditional diffusion model. Instead of relying on neural networks, the oracle provides accurate guidance in the form of estimated gradients, allowing the diffusion process to sample from a conditional distribution specified by quantum chemistry. We show that this results in more precise conditional generation of novel and stable molecular structures. Our experiments demonstrate that our method: (1) significantly reduces atomic forces, enhancing the validity of generated molecules when used for stability optimization; (2) is compatible with both explicit and implicit guidance in diffusion models, enabling joint optimization of molecular properties and stability; and (3) generalizes effectively to molecular optimization tasks beyond stability optimization.
Objective-Agnostic Enhancement of Molecule Properties via Multi-Stage VAE
Sep 10, 2023Abstract:Variational autoencoder (VAE) is a popular method for drug discovery and various architectures and pipelines have been proposed to improve its performance. However, VAE approaches are known to suffer from poor manifold recovery when the data lie on a low-dimensional manifold embedded in a higher dimensional ambient space [Dai and Wipf, 2019]. The consequences of it in drug discovery are somewhat under-explored. In this paper, we explore applying a multi-stage VAE approach, that can improve manifold recovery on a synthetic dataset, to the field of drug discovery. We experimentally evaluate our multi-stage VAE approach using the ChEMBL dataset and demonstrate its ability to improve the property statistics of generated molecules substantially from pre-existing methods without incorporating property predictors into the training pipeline. We further fine-tune our models on two curated and much smaller molecule datasets that target different proteins. Our experiments show an increase in the number of active molecules generated by the multi-stage VAE in comparison to their one-stage equivalent. For each of the two tasks, our baselines include methods that use learned property predictors to incorporate target metrics directly into the training objective and we discuss complications that arise with this methodology.
Improving Molecule Properties Through 2-Stage VAE
Dec 06, 2022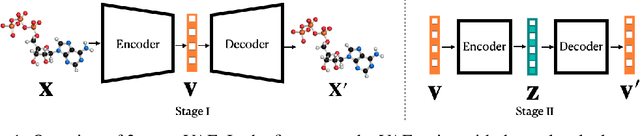
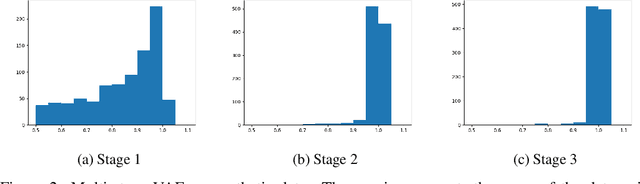
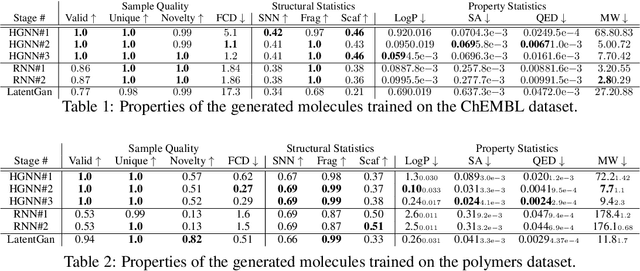
Abstract:Variational autoencoder (VAE) is a popular method for drug discovery and there had been a great deal of architectures and pipelines proposed to improve its performance. But the VAE model itself suffers from deficiencies such as poor manifold recovery when data lie on low-dimensional manifold embedded in higher dimensional ambient space and they manifest themselves in each applications differently. The consequences of it in drug discovery is somewhat under-explored. In this paper, we study how to improve the similarity of the data generated via VAE and the training dataset by improving manifold recovery via a 2-stage VAE where the second stage VAE is trained on the latent space of the first one. We experimentally evaluated our approach using the ChEMBL dataset as well as a polymer datasets. In both dataset, the 2-stage VAE method is able to improve the property statistics significantly from a pre-existing method.
Variational autoencoders in the presence of low-dimensional data: landscape and implicit bias
Dec 13, 2021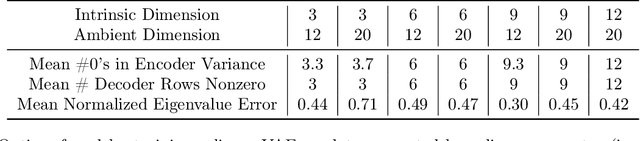
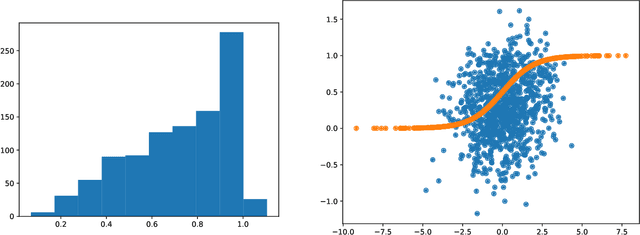
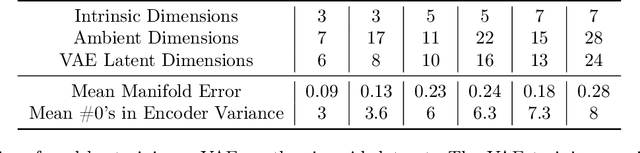
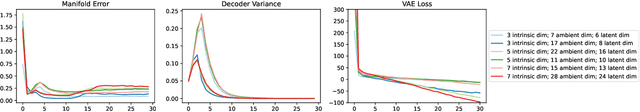
Abstract:Variational Autoencoders (VAEs) are one of the most commonly used generative models, particularly for image data. A prominent difficulty in training VAEs is data that is supported on a lower dimensional manifold. Recent work by Dai and Wipf (2019) suggests that on low-dimensional data, the generator will converge to a solution with 0 variance which is correctly supported on the ground truth manifold. In this paper, via a combination of theoretical and empirical results, we show that the story is more subtle. Precisely, we show that for linear encoders/decoders, the story is mostly true and VAE training does recover a generator with support equal to the ground truth manifold, but this is due to the implicit bias of gradient descent rather than merely the VAE loss itself. In the nonlinear case, we show that the VAE training frequently learns a higher-dimensional manifold which is a superset of the ground truth manifold.
Unsupervised Program Synthesis for Images using Tree-Structured LSTM
Jan 27, 2020

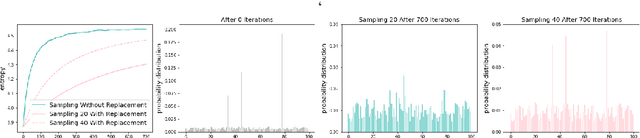

Abstract:Program synthesis has recently emerged as a promising approach to the image parsing task. However, most prior works have relied on supervised learning methods, which require ground truth programs for each training image. We present an unsupervised learning algorithm that can parse constructive solid geometry (CSG) images into context-free grammar with a non-differentiable renderer. We propose a grammar-encoded tree LSTM to effectively constrain our search space by leveraging the structure of the context-free grammar while handling the non-differentiable renderer via REINFORCE and encouraging the exploration by regularizing the objective with an entropy term. Instead of using simple Monte Carlo sampling, we propose a lower-variance entropy estimator with sampling without replacement for effective exploration. We demonstrate the effectiveness of the proposed algorithm on a synthetic 2D CSG dataset, which outperforms baseline models by a large margin.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge