Improving Molecule Properties Through 2-Stage VAE
Paper and Code
Dec 06, 2022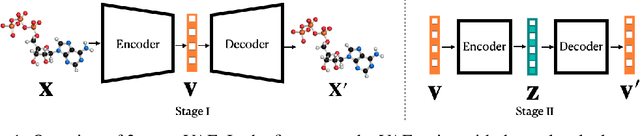
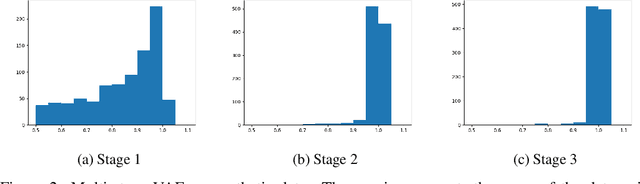
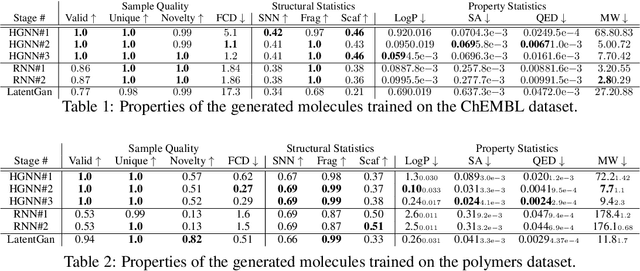
Variational autoencoder (VAE) is a popular method for drug discovery and there had been a great deal of architectures and pipelines proposed to improve its performance. But the VAE model itself suffers from deficiencies such as poor manifold recovery when data lie on low-dimensional manifold embedded in higher dimensional ambient space and they manifest themselves in each applications differently. The consequences of it in drug discovery is somewhat under-explored. In this paper, we study how to improve the similarity of the data generated via VAE and the training dataset by improving manifold recovery via a 2-stage VAE where the second stage VAE is trained on the latent space of the first one. We experimentally evaluated our approach using the ChEMBL dataset as well as a polymer datasets. In both dataset, the 2-stage VAE method is able to improve the property statistics significantly from a pre-existing method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge