Chaitanya Dwivedi
Performance Prediction of Hub-Based Swarms
Aug 09, 2024Abstract:A hub-based colony consists of multiple agents who share a common nest site called the hub. Agents perform tasks away from the hub like foraging for food or gathering information about future nest sites. Modeling hub-based colonies is challenging because the size of the collective state space grows rapidly as the number of agents grows. This paper presents a graph-based representation of the colony that can be combined with graph-based encoders to create low-dimensional representations of collective state that can scale to many agents for a best-of-N colony problem. We demonstrate how the information in the low-dimensional embedding can be used with two experiments. First, we show how the information in the tensor can be used to cluster collective states by the probability of choosing the best site for a very small problem. Second, we show how structured collective trajectories emerge when a graph encoder is used to learn the low-dimensional embedding, and these trajectories have information that can be used to predict swarm performance.
Multi stain graph fusion for multimodal integration in pathology
Apr 26, 2022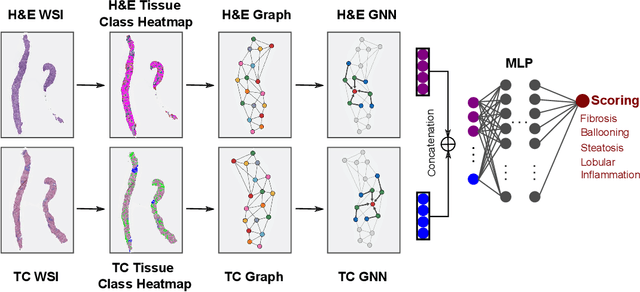
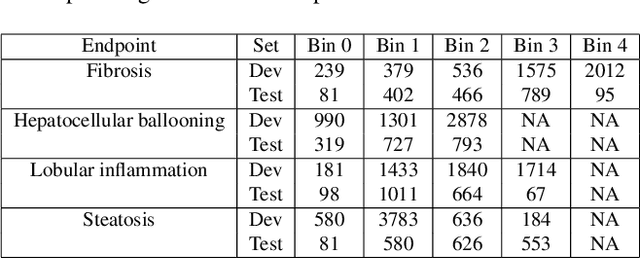
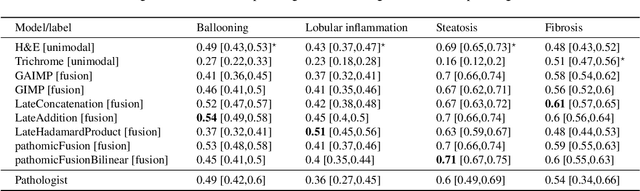
Abstract:In pathology, tissue samples are assessed using multiple staining techniques to enhance contrast in unique histologic features. In this paper, we introduce a multimodal CNN-GNN based graph fusion approach that leverages complementary information from multiple non-registered histopathology images to predict pathologic scores. We demonstrate this approach in nonalcoholic steatohepatitis (NASH) by predicting CRN fibrosis stage and NAFLD Activity Score (NAS). Primary assessment of NASH typically requires liver biopsy evaluation on two histological stains: Trichrome (TC) and hematoxylin and eosin (H&E). Our multimodal approach learns to extract complementary information from TC and H&E graphs corresponding to each stain while simultaneously learning an optimal policy to combine this information. We report up to 20% improvement in predicting fibrosis stage and NAS component grades over single-stain modeling approaches, measured by computing linearly weighted Cohen's kappa between machine-derived vs. pathologist consensus scores. Broadly, this paper demonstrates the value of leveraging diverse pathology images for improved ML-powered histologic assessment.
Probabilistic Neighbourhood Component Analysis: Sample Efficient Uncertainty Estimation in Deep Learning
Jul 18, 2020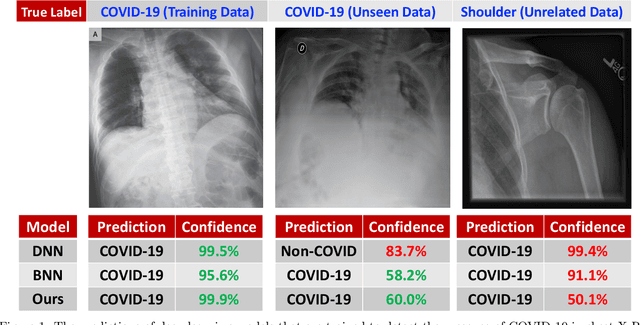
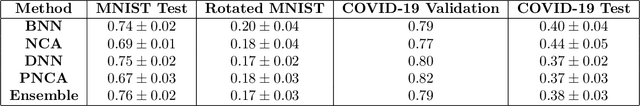
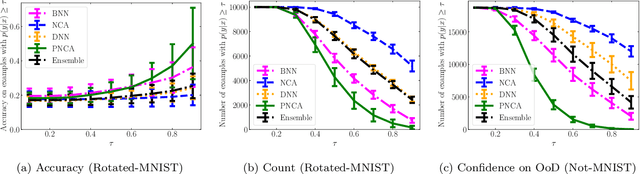
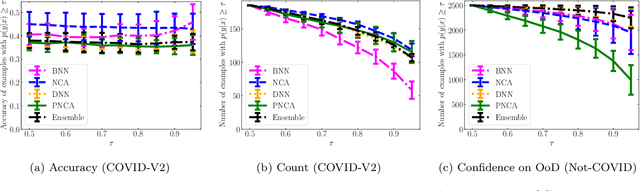
Abstract:While Deep Neural Networks (DNNs) achieve state-of-the-art accuracy in various applications, they often fall short in accurately estimating their predictive uncertainty and, in turn, fail to recognize when these predictions may be wrong. Several uncertainty-aware models, such as Bayesian Neural Network (BNNs) and Deep Ensembles have been proposed in the literature for quantifying predictive uncertainty. However, research in this area has been largely confined to the big data regime. In this work, we show that the uncertainty estimation capability of state-of-the-art BNNs and Deep Ensemble models degrades significantly when the amount of training data is small. To address the issue of accurate uncertainty estimation in the small-data regime, we propose a probabilistic generalization of the popular sample-efficient non-parametric kNN approach. Our approach enables deep kNN classifier to accurately quantify underlying uncertainties in its prediction. We demonstrate the usefulness of the proposed approach by achieving superior uncertainty quantification as compared to state-of-the-art on a real-world application of COVID-19 diagnosis from chest X-Rays. Our code is available at https://github.com/ankurmallick/sample-efficient-uq
Deep Probabilistic Kernels for Sample-Efficient Learning
Oct 13, 2019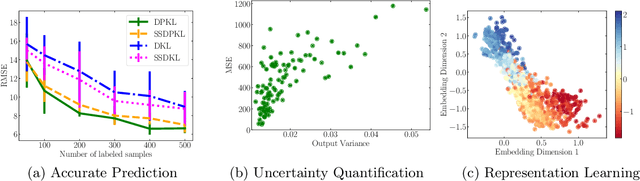
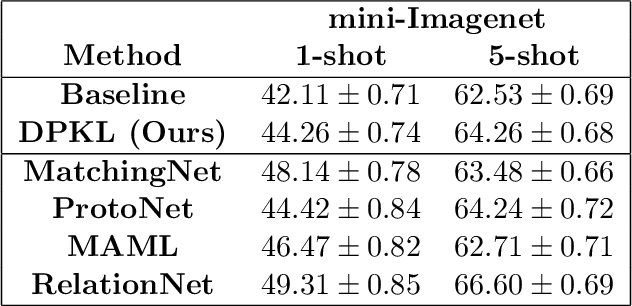
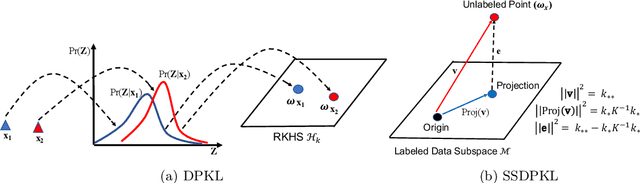
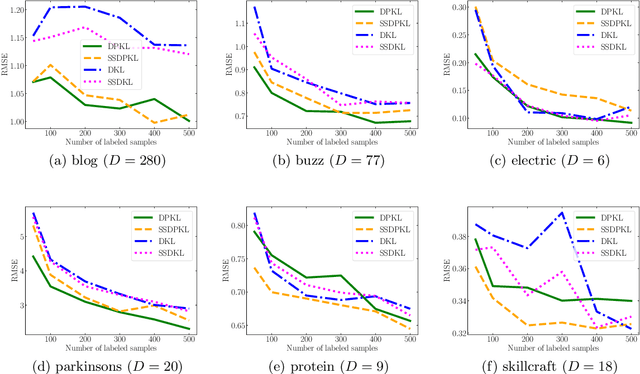
Abstract:Gaussian Processes (GPs) with an appropriate kernel are known to provide accurate predictions and uncertainty estimates even with very small amounts of labeled data. However, GPs are generally unable to learn a good representation that can encode intricate structures in high dimensional data. The representation power of GPs depends heavily on kernel functions used to quantify the similarity between data points. Traditional GP kernels are not very effective at capturing similarity between high dimensional data points, while methods that use deep neural networks to learn a kernel are not sample-efficient. To overcome these drawbacks, we propose deep probabilistic kernels which use a probabilistic neural network to map high-dimensional data to a probability distribution in a low dimensional subspace, and leverage the rich work on kernels between distributions to capture the similarity between these distributions. Experiments on a variety of datasets show that building a GP using this covariance kernel solves the conflicting problems of representation learning and sample efficiency. Our model can be extended beyond GPs to other small-data paradigms such as few-shot classification where we show competitive performance with state-of-the-art models on the mini-Imagenet dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge