Chae Jung Park
Department of Radiology, Yongin Severance Hospital, Yonsei University College of Medicine, Yongin, Republic of Korea
Symmetric masking strategy enhances the performance of Masked Image Modeling
Aug 23, 2024Abstract:Masked Image Modeling (MIM) is a technique in self-supervised learning that focuses on acquiring detailed visual representations from unlabeled images by estimating the missing pixels in randomly masked sections. It has proven to be a powerful tool for the preliminary training of Vision Transformers (ViTs), yielding impressive results across various tasks. Nevertheless, most MIM methods heavily depend on the random masking strategy to formulate the pretext task. This strategy necessitates numerous trials to ascertain the optimal dropping ratio, which can be resource-intensive, requiring the model to be pre-trained for anywhere between 800 to 1600 epochs. Furthermore, this approach may not be suitable for all datasets. In this work, we propose a new masking strategy that effectively helps the model capture global and local features. Based on this masking strategy, SymMIM, our proposed training pipeline for MIM is introduced. SymMIM achieves a new SOTA accuracy of 85.9\% on ImageNet using ViT-Large and surpasses previous SOTA across downstream tasks such as image classification, semantic segmentation, object detection, instance segmentation tasks, and so on.
SAVE: Segment Audio-Visual Easy way using Segment Anything Model
Jul 02, 2024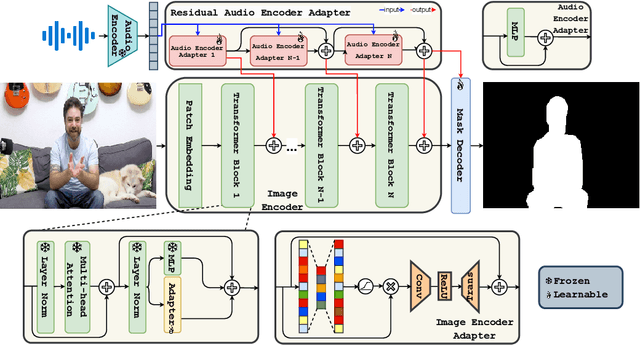
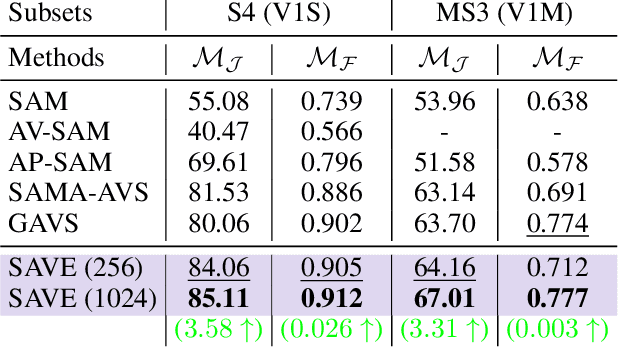
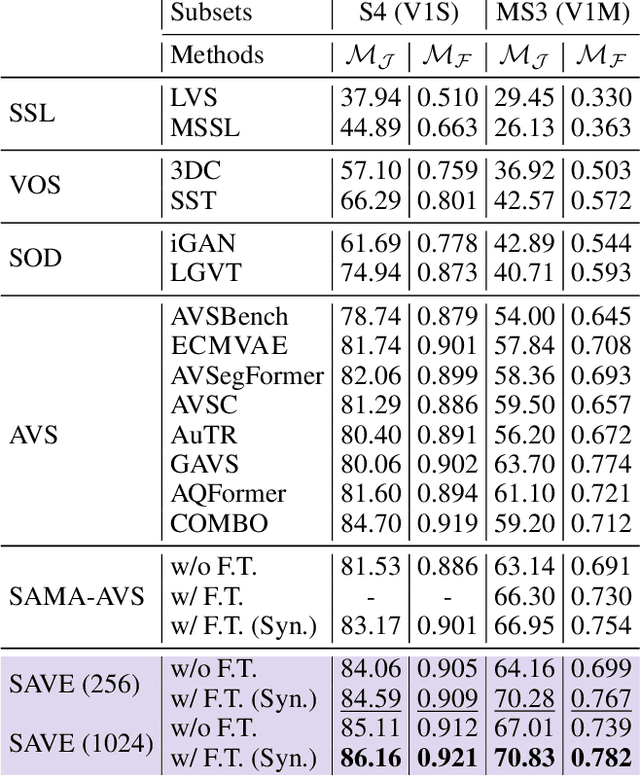

Abstract:The primary aim of Audio-Visual Segmentation (AVS) is to precisely identify and locate auditory elements within visual scenes by accurately predicting segmentation masks at the pixel level. Achieving this involves comprehensively considering data and model aspects to address this task effectively. This study presents a lightweight approach, SAVE, which efficiently adapts the pre-trained segment anything model (SAM) to the AVS task. By incorporating an image encoder adapter into the transformer blocks to better capture the distinct dataset information and proposing a residual audio encoder adapter to encode the audio features as a sparse prompt, our proposed model achieves effective audio-visual fusion and interaction during the encoding stage. Our proposed method accelerates the training and inference speed by reducing the input resolution from 1024 to 256 pixels while achieving higher performance compared with the previous SOTA. Extensive experimentation validates our approach, demonstrating that our proposed model outperforms other SOTA methods significantly. Moreover, leveraging the pre-trained model on synthetic data enhances performance on real AVSBench data, achieving 84.59 mIoU on the S4 (V1S) subset and 70.28 mIoU on the MS3 (V1M) set with only 256 pixels for input images. This increases up to 86.16 mIoU on the S4 (V1S) and 70.83 mIoU on the MS3 (V1M) with inputs of 1024 pixels.
Retro: Reusing teacher projection head for efficient embedding distillation on Lightweight Models via Self-supervised Learning
May 27, 2024Abstract:Self-supervised learning (SSL) is gaining attention for its ability to learn effective representations with large amounts of unlabeled data. Lightweight models can be distilled from larger self-supervised pre-trained models using contrastive and consistency constraints. Still, the different sizes of the projection heads make it challenging for students to mimic the teacher's embedding accurately. We propose \textsc{Retro}, which reuses the teacher's projection head for students, and our experimental results demonstrate significant improvements over the state-of-the-art on all lightweight models. For instance, when training EfficientNet-B0 using ResNet-50/101/152 as teachers, our approach improves the linear result on ImageNet to $66.9\%$, $69.3\%$, and $69.8\%$, respectively, with significantly fewer parameters.
A human brain atlas of chi-separation for normative iron and myelin distributions
Nov 08, 2023Abstract:Iron and myelin are primary susceptibility sources in the human brain. These substances are essential for healthy brain, and their abnormalities are often related to various neurological disorders. Recently, an advanced susceptibility mapping technique, which is referred to as chi-separation, has been proposed successfully disentangling paramagnetic iron from diamagnetic myelin, opening a new potential for generating iron map and myelin map in the brain. Utilizing this technique, this study constructs a normative chi-separation atlas from 106 healthy human brains. The resulting atlas provides detailed anatomical structures associated with the distributions of iron and myelin, clearly delineating subcortical nuclei and white matter fiber bundles. Additionally, susceptibility values in a number of regions of interest are reported along with age-dependent changes. This atlas may have direct applications such as localization of subcortical structures for deep brain stimulation or high-intensity focused ultrasound and also serve as a valuable resource for future research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge