Carlos Outeiral
Generative artificial intelligence for de novo protein design
Oct 15, 2023Abstract:Engineering new molecules with desirable functions and properties has the potential to extend our ability to engineer proteins beyond what nature has so far evolved. Advances in the so-called "de novo" design problem have recently been brought forward by developments in artificial intelligence. Generative architectures, such as language models and diffusion processes, seem adept at generating novel, yet realistic proteins that display desirable properties and perform specified functions. State-of-the-art design protocols now achieve experimental success rates nearing 20%, thus widening the access to de novo designed proteins. Despite extensive progress, there are clear field-wide challenges, for example in determining the best in silico metrics to prioritise designs for experimental testing, and in designing proteins that can undergo large conformational changes or be regulated by post-translational modifications and other cellular processes. With an increase in the number of models being developed, this review provides a framework to understand how these tools fit into the overall process of de novo protein design. Throughout, we highlight the power of incorporating biochemical knowledge to improve performance and interpretability.
The prospects of quantum computing in computational molecular biology
May 26, 2020
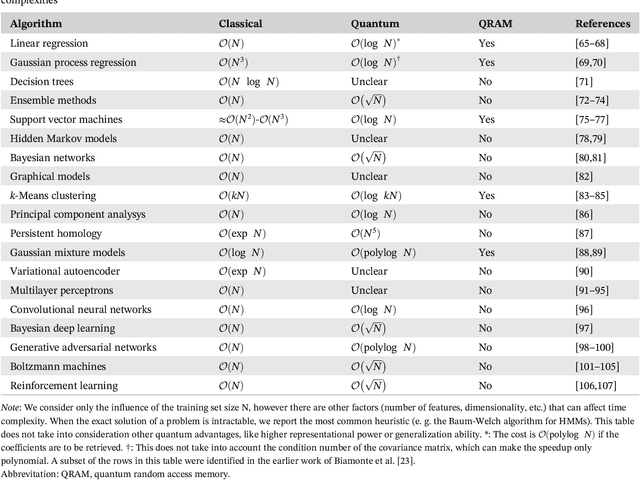
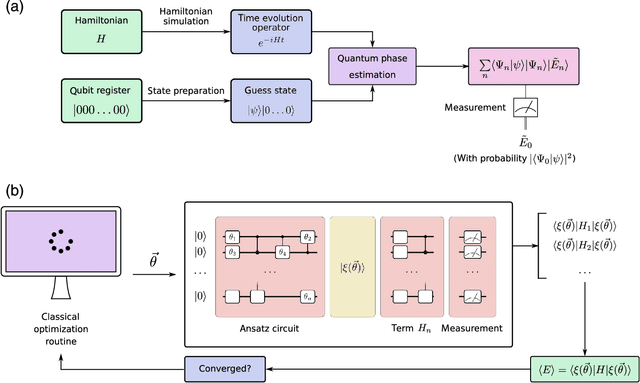

Abstract:Quantum computers can in principle solve certain problems exponentially more quickly than their classical counterparts. We have not yet reached the advent of useful quantum computation, but when we do, it will affect nearly all scientific disciplines. In this review, we examine how current quantum algorithms could revolutionize computational biology and bioinformatics. There are potential benefits across the entire field, from the ability to process vast amounts of information and run machine learning algorithms far more efficiently, to algorithms for quantum simulation that are poised to improve computational calculations in drug discovery, to quantum algorithms for optimization that may advance fields from protein structure prediction to network analysis. However, these exciting prospects are susceptible to "hype", and it is also important to recognize the caveats and challenges in this new technology. Our aim is to introduce the promise and limitations of emerging quantum computing technologies in the areas of computational molecular biology and bioinformatics.
* 23 pages, 3 figures
Objective-Reinforced Generative Adversarial Networks (ORGAN) for Sequence Generation Models
Feb 07, 2018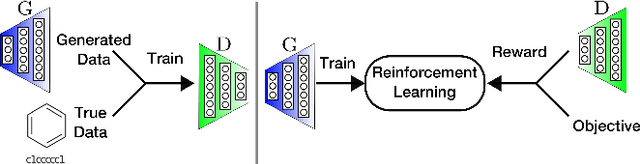



Abstract:In unsupervised data generation tasks, besides the generation of a sample based on previous observations, one would often like to give hints to the model in order to bias the generation towards desirable metrics. We propose a method that combines Generative Adversarial Networks (GANs) and reinforcement learning (RL) in order to accomplish exactly that. While RL biases the data generation process towards arbitrary metrics, the GAN component of the reward function ensures that the model still remembers information learned from data. We build upon previous results that incorporated GANs and RL in order to generate sequence data and test this model in several settings for the generation of molecules encoded as text sequences (SMILES) and in the context of music generation, showing for each case that we can effectively bias the generation process towards desired metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge