Carlos G. Oliver
Edge-similarity-aware Graph Neural Networks
Sep 20, 2021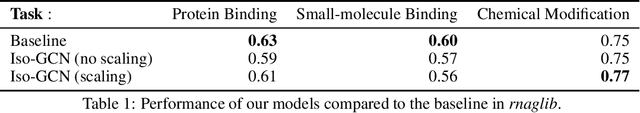
Abstract:Graph are a ubiquitous data representation, as they represent a flexible and compact representation. For instance, the 3D structure of RNA can be efficiently represented as $\textit{2.5D graphs}$, graphs whose nodes are nucleotides and edges represent chemical interactions. In this setting, we have biological evidence of the similarity between the edge types, as some chemical interactions are more similar than others. Machine learning on graphs have recently experienced a breakthrough with the introduction of Graph Neural Networks. This algorithm can be framed as a message passing algorithm between graph nodes over graph edges. These messages can depend on the edge type they are transmitted through, but no method currently constrains how a message is altered when the edge type changes. Motivated by the RNA use case, in this project we introduce a graph neural network layer which can leverage prior information about similarities between edges. We show that despite the theoretical appeal of including this similarity prior, the empirical performance is not enhanced on the tasks and datasets we include here.
Leveraging binding-site structure for drug discovery with point-cloud methods
May 28, 2019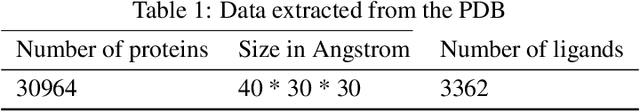


Abstract:Computational drug discovery strategies can be broadly placed in two categories: ligand-based methods which identify novel molecules by similarity with known ligands, and structure-based methods which predict molecules with high-affinity to a given 3D structure (e.g. a protein). However, ligand-based methods do not leverage information about the binding site, and structure-based approaches rely on the knowledge of a finite set of ligands binding the target. In this work, we introduce TarLig, a novel approach that aims to bridge the gap between ligand and structure-based approaches. We use the 3D structure of the binding site as input to a model which predicts the ligand preferences of the binding site. The resulting predictions could then offer promising seeds and constraints in the chemical space search, based on the binding site structure. TarLig outperforms standard models by introducing a data-alignment and augmentation technique. The recent popularity of Volumetric 3DCNN pipelines in structural bioinformatics suggests that this extra step could help a wide range of methods to improve their results with minimal modifications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge