Edge-similarity-aware Graph Neural Networks
Paper and Code
Sep 20, 2021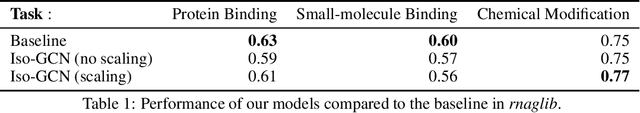
Graph are a ubiquitous data representation, as they represent a flexible and compact representation. For instance, the 3D structure of RNA can be efficiently represented as $\textit{2.5D graphs}$, graphs whose nodes are nucleotides and edges represent chemical interactions. In this setting, we have biological evidence of the similarity between the edge types, as some chemical interactions are more similar than others. Machine learning on graphs have recently experienced a breakthrough with the introduction of Graph Neural Networks. This algorithm can be framed as a message passing algorithm between graph nodes over graph edges. These messages can depend on the edge type they are transmitted through, but no method currently constrains how a message is altered when the edge type changes. Motivated by the RNA use case, in this project we introduce a graph neural network layer which can leverage prior information about similarities between edges. We show that despite the theoretical appeal of including this similarity prior, the empirical performance is not enhanced on the tasks and datasets we include here.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge