Carl Nettelblad
Learning Genomic Structure from $k$-mers
May 22, 2025Abstract:Sequencing a genome to determine an individual's DNA produces an enormous number of short nucleotide subsequences known as reads, which must be reassembled to reconstruct the full genome. We present a method for analyzing this type of data using contrastive learning, in which an encoder model is trained to produce embeddings that cluster together sequences from the same genomic region. The sequential nature of genomic regions is preserved in the form of trajectories through this embedding space. Trained solely to reflect the structure of the genome, the resulting model provides a general representation of $k$-mer sequences, suitable for a range of downstream tasks involving read data. We apply our framework to learn the structure of the $E.\ coli$ genome, and demonstrate its use in simulated ancient DNA (aDNA) read mapping and identification of structural variations. Furthermore, we illustrate the potential of using this type of model for metagenomic species identification. We show how incorporating a domain-specific noise model can enhance embedding robustness, and how a supervised contrastive learning setting can be adopted when a linear reference genome is available, by introducing a distance thresholding parameter $\Gamma$. The model can also be trained fully self-supervised on read data, enabling analysis without the need to construct a full genome assembly using specialized algorithms. Small prediction heads based on a pre-trained embedding are shown to perform on par with BWA-aln, the current gold standard approach for aDNA mapping, in terms of accuracy and runtime for short genomes. Given the method's favorable scaling properties with respect to total genome size, inference using our approach is highly promising for metagenomic applications and for mapping to genomes comparable in size to the human genome.
Training Algorithm Matters for the Performance of Neural Network Potential
Sep 08, 2021



Abstract:One hidden yet important issue for developing neural network potentials (NNPs) is the choice of training algorithm. Here we compare the performance of two popular training algorithms, the adaptive moment estimation algorithm (Adam) and the extended Kalman filter algorithm (EKF), using the Behler-Parrinello neural network (BPNN) and two publicly accessible datasets of liquid water. It is found that NNPs trained with EKF are more transferable and less sensitive to the value of the learning rate, as compared to Adam. In both cases, error metrics of the test set do not always serve as a good indicator for the actual performance of NNPs. Instead, we show that their performance correlates well with a Fisher information based similarity measure.
Bootstrapping Weakly Supervised Segmentation-free Word Spotting through HMM-based Alignment
Mar 24, 2020


Abstract:Recent work in word spotting in handwritten documents has yielded impressive results. This progress has largely been made by supervised learning systems, which are dependent on manually annotated data, making deployment to new collections a significant effort. In this paper, we propose an approach that utilises transcripts without bounding box annotations to train segmentation-free query-by-string word spotting models, given a partially trained model. This is done through a training-free alignment procedure based on hidden Markov models. This procedure creates a tentative mapping between word region proposals and the transcriptions to automatically create additional weakly annotated training data, without choosing any single alignment possibility as the correct one. When only using between 1% and 7% of the fully annotated training sets for partial convergence, we automatically annotate the remaining training data and successfully train using it. On all our datasets, our final trained model then comes within a few mAP% of the performance from a model trained with the full training set used as ground truth. We believe that this will be a significant advance towards a more general use of word spotting, since digital transcription data will already exist for parts of many collections of interest.
Flash X-ray diffraction imaging in 3D: a proposed analysis pipeline
Oct 30, 2019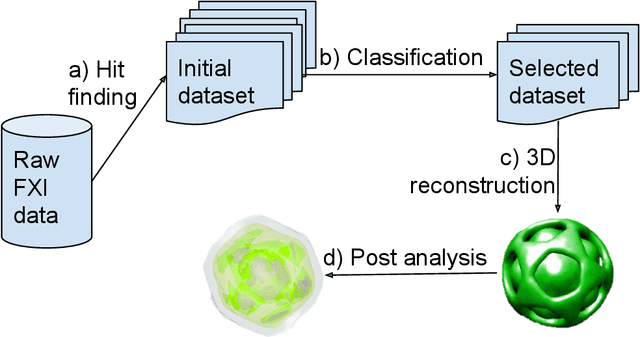
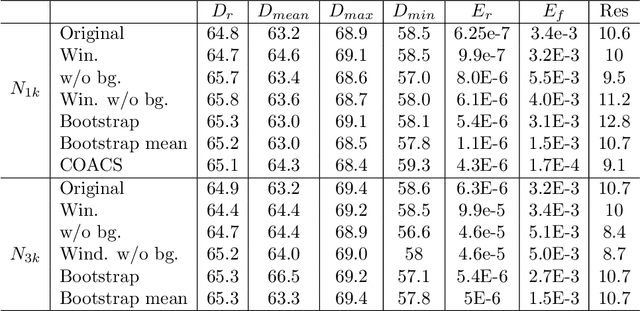
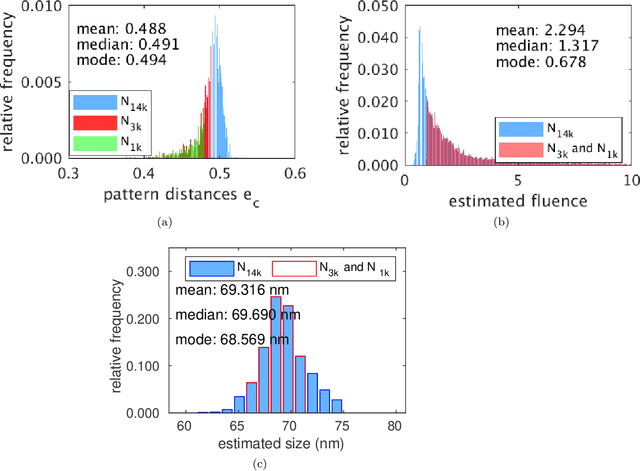
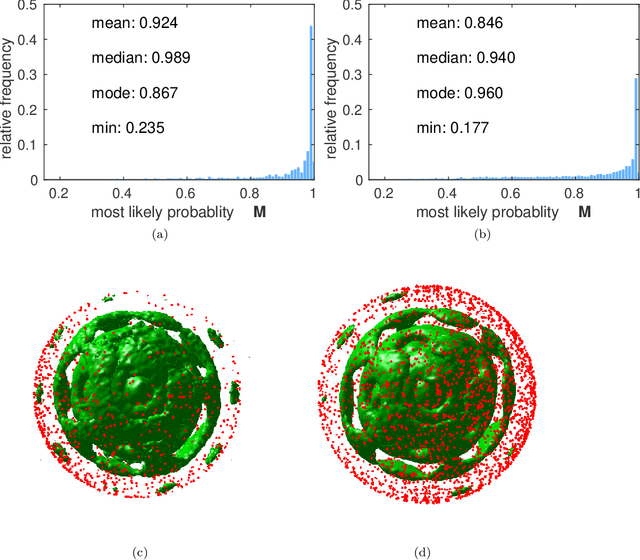
Abstract:Modern Flash X-ray diffraction imaging (FXI) acquires diffraction signals from single biomolecules at a high repetition rate from Free X-ray Electron lasers (XFELs), easily obtaining millions of 2D diffraction patterns from a single experiment. Due to the stochastic nature of FXI experiments and the massive volumes of data, retrieving 3D electron densities from raw 2D diffraction patterns is a challenging and time-consuming task. We propose a semi-automatic data analysis pipeline for FXI experiments, which includes four steps: hit finding and preliminary filtering, pattern classification, 3D Fourier reconstruction, and post analysis. We also include a recently developed bootstrap methodology in the post-analysis step for uncertainty analysis and quality control. To achieve the best possible resolution, we further suggest using background subtraction, signal windowing, and convex optimization techniques when retrieving the Fourier phases in the post-analysis step. As an application example, we quantified the 3D electron structure of the PR772 virus using the proposed data-analysis pipeline. The retrieved structure was above the detector-edge resolution and clearly showed the pseudo-icosahedral capsid of the PR772.
Assessing Uncertainties in X-ray Single-particle Three-dimensional reconstructions
Jan 02, 2017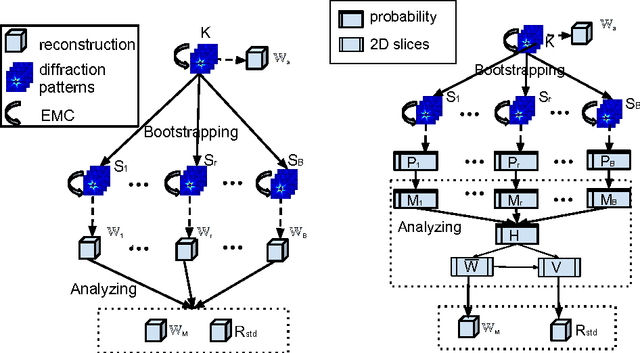
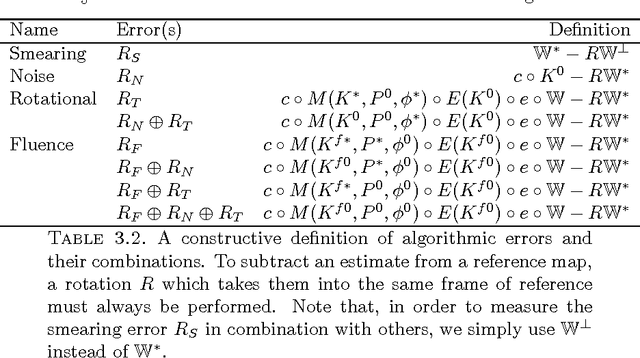
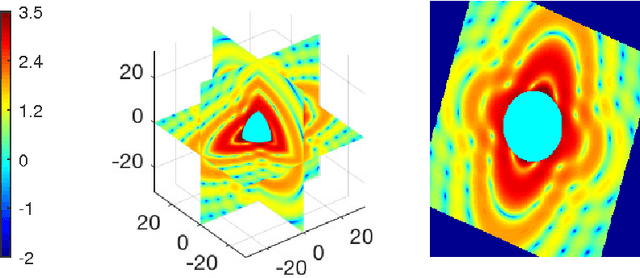
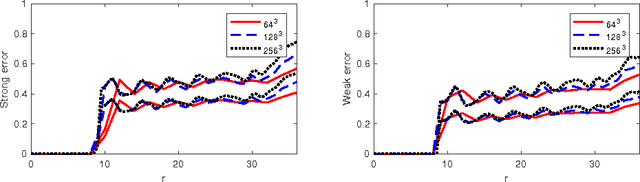
Abstract:Modern technology for producing extremely bright and coherent X-ray laser pulses provides the possibility to acquire a large number of diffraction patterns from individual biological nanoparticles, including proteins, viruses, and DNA. These two-dimensional diffraction patterns can be practically reconstructed and retrieved down to a resolution of a few \angstrom. In principle, a sufficiently large collection of diffraction patterns will contain the required information for a full three-dimensional reconstruction of the biomolecule. The computational methodology for this reconstruction task is still under development and highly resolved reconstructions have not yet been produced. We analyze the Expansion-Maximization-Compression scheme, the current state of the art approach for this very challenging application, by isolating different sources of uncertainty. Through numerical experiments on synthetic data we evaluate their respective impact. We reach conclusions of relevance for handling actual experimental data, as well as pointing out certain improvements to the underlying estimation algorithm. We also introduce a practically applicable computational methodology in the form of bootstrap procedures for assessing reconstruction uncertainty in the real data case. We evaluate the sharpness of this approach and argue that this type of procedure will be critical in the near future when handling the increasing amount of data.
* 21 pages
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge