Bora Kargi
Scholar Inbox: Personalized Paper Recommendations for Scientists
Apr 11, 2025
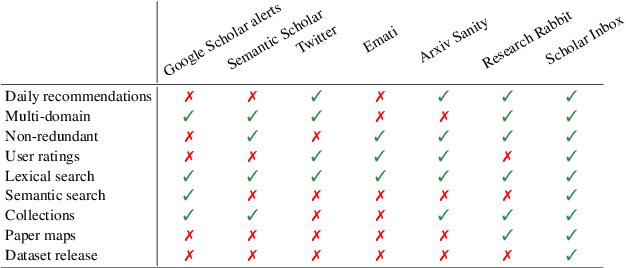

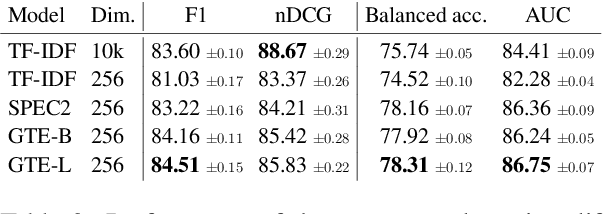
Abstract:Scholar Inbox is a new open-access platform designed to address the challenges researchers face in staying current with the rapidly expanding volume of scientific literature. We provide personalized recommendations, continuous updates from open-access archives (arXiv, bioRxiv, etc.), visual paper summaries, semantic search, and a range of tools to streamline research workflows and promote open research access. The platform's personalized recommendation system is trained on user ratings, ensuring that recommendations are tailored to individual researchers' interests. To further enhance the user experience, Scholar Inbox also offers a map of science that provides an overview of research across domains, enabling users to easily explore specific topics. We use this map to address the cold start problem common in recommender systems, as well as an active learning strategy that iteratively prompts users to rate a selection of papers, allowing the system to learn user preferences quickly. We evaluate the quality of our recommendation system on a novel dataset of 800k user ratings, which we make publicly available, as well as via an extensive user study. https://www.scholar-inbox.com/
Evaluating Self-Supervised Learning in Medical Imaging: A Benchmark for Robustness, Generalizability, and Multi-Domain Impact
Dec 26, 2024Abstract:Self-supervised learning (SSL) has emerged as a promising paradigm in medical imaging, addressing the chronic challenge of limited labeled data in healthcare settings. While SSL has shown impressive results, existing studies in the medical domain are often limited in scope, focusing on specific datasets or modalities, or evaluating only isolated aspects of model performance. This fragmented evaluation approach poses a significant challenge, as models deployed in critical medical settings must not only achieve high accuracy but also demonstrate robust performance and generalizability across diverse datasets and varying conditions. To address this gap, we present a comprehensive evaluation of SSL methods within the medical domain, with a particular focus on robustness and generalizability. Using the MedMNIST dataset collection as a standardized benchmark, we evaluate 8 major SSL methods across 11 different medical datasets. Our study provides an in-depth analysis of model performance in both in-domain scenarios and the detection of out-of-distribution (OOD) samples, while exploring the effect of various initialization strategies, model architectures, and multi-domain pre-training. We further assess the generalizability of SSL methods through cross-dataset evaluations and the in-domain performance with varying label proportions (1%, 10%, and 100%) to simulate real-world scenarios with limited supervision. We hope this comprehensive benchmark helps practitioners and researchers make more informed decisions when applying SSL methods to medical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge