Bindu Nanduri
Bayesian-Guided Generation of Synthetic Microbiomes with Minimized Pathogenicity
Apr 29, 2024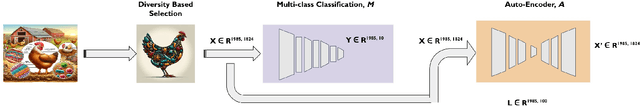
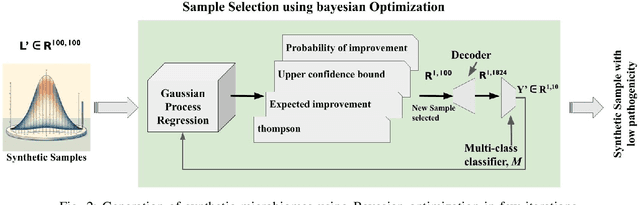
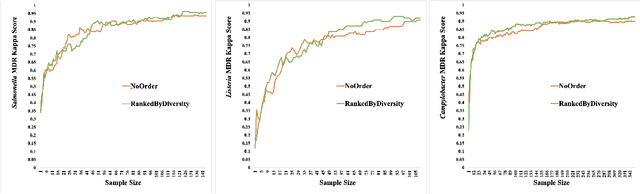
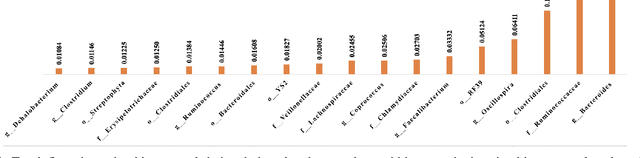
Abstract:Synthetic microbiomes offer new possibilities for modulating microbiota, to address the barriers in multidtug resistance (MDR) research. We present a Bayesian optimization approach to enable efficient searching over the space of synthetic microbiome variants to identify candidates predictive of reduced MDR. Microbiome datasets were encoded into a low-dimensional latent space using autoencoders. Sampling from this space allowed generation of synthetic microbiome signatures. Bayesian optimization was then implemented to select variants for biological screening to maximize identification of designs with restricted MDR pathogens based on minimal samples. Four acquisition functions were evaluated: expected improvement, upper confidence bound, Thompson sampling, and probability of improvement. Based on each strategy, synthetic samples were prioritized according to their MDR detection. Expected improvement, upper confidence bound, and probability of improvement consistently produced synthetic microbiome candidates with significantly fewer searches than Thompson sampling. By combining deep latent space mapping and Bayesian learning for efficient guided screening, this study demonstrated the feasibility of creating bespoke synthetic microbiomes with customized MDR profiles.
EndToEndML: An Open-Source End-to-End Pipeline for Machine Learning Applications
Mar 27, 2024



Abstract:Artificial intelligence (AI) techniques are widely applied in the life sciences. However, applying innovative AI techniques to understand and deconvolute biological complexity is hindered by the learning curve for life science scientists to understand and use computing languages. An open-source, user-friendly interface for AI models, that does not require programming skills to analyze complex biological data will be extremely valuable to the bioinformatics community. With easy access to different sequencing technologies and increased interest in different 'omics' studies, the number of biological datasets being generated has increased and analyzing these high-throughput datasets is computationally demanding. The majority of AI libraries today require advanced programming skills as well as machine learning, data preprocessing, and visualization skills. In this research, we propose a web-based end-to-end pipeline that is capable of preprocessing, training, evaluating, and visualizing machine learning (ML) models without manual intervention or coding expertise. By integrating traditional machine learning and deep neural network models with visualizations, our library assists in recognizing, classifying, clustering, and predicting a wide range of multi-modal, multi-sensor datasets, including images, languages, and one-dimensional numerical data, for drug discovery, pathogen classification, and medical diagnostics.
Deep Sensitivity Analysis for Objective-Oriented Combinatorial Optimization
Feb 28, 2024Abstract:Pathogen control is a critical aspect of modern poultry farming, providing important benefits for both public health and productivity. Effective poultry management measures to reduce pathogen levels in poultry flocks promote food safety by lowering risks of food-borne illnesses. They also support animal health and welfare by preventing infectious diseases that can rapidly spread and impact flock growth, egg production, and overall health. This study frames the search for optimal management practices that minimize the presence of multiple pathogens as a combinatorial optimization problem. Specifically, we model the various possible combinations of management settings as a solution space that can be efficiently explored to identify configurations that optimally reduce pathogen levels. This design incorporates a neural network feedback-based method that combines feature explanations with global sensitivity analysis to ensure combinatorial optimization in multiobjective settings. Our preliminary experiments have promising results when applied to two real-world agricultural datasets. While further validation is still needed, these early experimental findings demonstrate the potential of the model to derive targeted feature interactions that adaptively optimize pathogen control under varying real-world constraints.
Towards Interpreting Multi-Objective Feature Associations
Feb 28, 2024Abstract:Understanding how multiple features are associated and contribute to a specific objective is as important as understanding how each feature contributes to a particular outcome. Interpretability of a single feature in a prediction may be handled in multiple ways; however, in a multi-objective prediction, it is difficult to obtain interpretability of a combination of feature values. To address this issue, we propose an objective specific feature interaction design using multi-labels to find the optimal combination of features in agricultural settings. One of the novel aspects of this design is the identification of a method that integrates feature explanations with global sensitivity analysis in order to ensure combinatorial optimization in multi-objective settings. We have demonstrated in our preliminary experiments that an approximate combination of feature values can be found to achieve the desired outcome using two agricultural datasets: one with pre-harvest poultry farm practices for multi-drug resistance presence, and one with post-harvest poultry farm practices for food-borne pathogens. In our combinatorial optimization approach, all three pathogens are taken into consideration simultaneously to account for the interaction between conditions that favor different types of pathogen growth. These results indicate that explanation-based approaches are capable of identifying combinations of features that reduce pathogen presence in fewer iterations than a baseline.
Information-theoretic Interestingness Measures for Cross-Ontology Data Mining
May 16, 2016



Abstract:Community annotation of biological entities with concepts from multiple bio-ontologies has created large and growing repositories of ontology-based annotation data with embedded implicit relationships among orthogonal ontologies. Development of efficient data mining methods and metrics to mine and assess the quality of the mined relationships has not kept pace with the growth of annotation data. In this study, we present a data mining method that uses ontology-guided generalization to discover relationships across ontologies along with a new interestingness metric based on information theory. We apply our data mining algorithm and interestingness measures to datasets from the Gene Expression Database at the Mouse Genome Informatics as a preliminary proof of concept to mine relationships between developmental stages in the mouse anatomy ontology and Gene Ontology concepts (biological process, molecular function and cellular component). In addition, we present a comparison of our interestingness metric to four existing metrics. Ontology-based annotation datasets provide a valuable resource for discovery of relationships across ontologies. The use of efficient data mining methods and appropriate interestingness metrics enables the identification of high quality relationships.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge