Michael J Rothrock Jr.
Bayesian-Guided Generation of Synthetic Microbiomes with Minimized Pathogenicity
Apr 29, 2024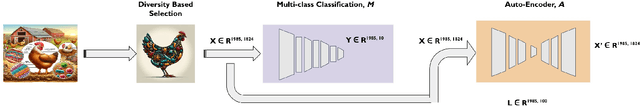
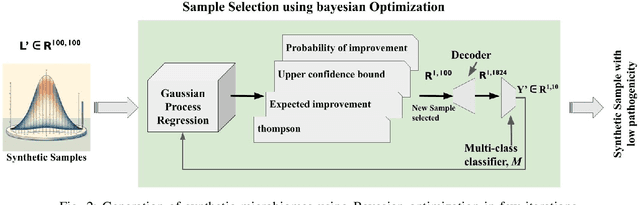
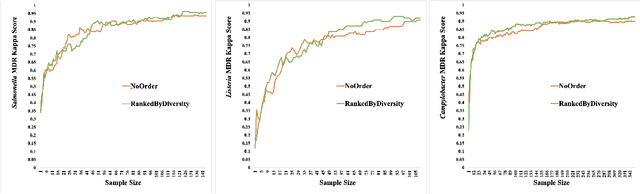
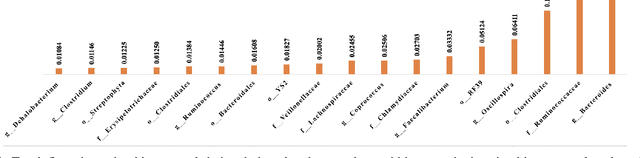
Abstract:Synthetic microbiomes offer new possibilities for modulating microbiota, to address the barriers in multidtug resistance (MDR) research. We present a Bayesian optimization approach to enable efficient searching over the space of synthetic microbiome variants to identify candidates predictive of reduced MDR. Microbiome datasets were encoded into a low-dimensional latent space using autoencoders. Sampling from this space allowed generation of synthetic microbiome signatures. Bayesian optimization was then implemented to select variants for biological screening to maximize identification of designs with restricted MDR pathogens based on minimal samples. Four acquisition functions were evaluated: expected improvement, upper confidence bound, Thompson sampling, and probability of improvement. Based on each strategy, synthetic samples were prioritized according to their MDR detection. Expected improvement, upper confidence bound, and probability of improvement consistently produced synthetic microbiome candidates with significantly fewer searches than Thompson sampling. By combining deep latent space mapping and Bayesian learning for efficient guided screening, this study demonstrated the feasibility of creating bespoke synthetic microbiomes with customized MDR profiles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge