Artur Lichtenberg
Impact of Leakage on Data Harmonization in Machine Learning Pipelines in Class Imbalance Across Sites
Oct 25, 2024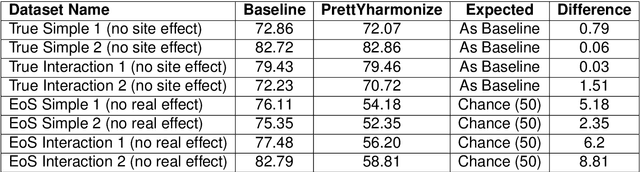
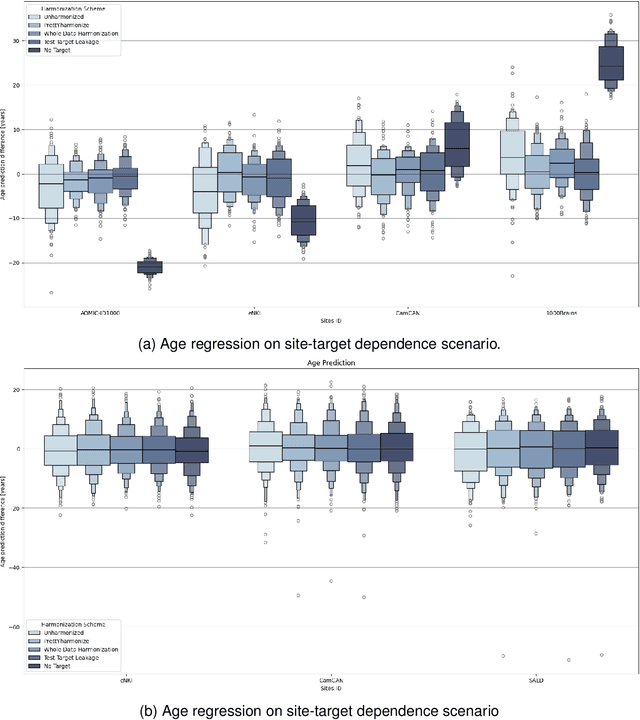
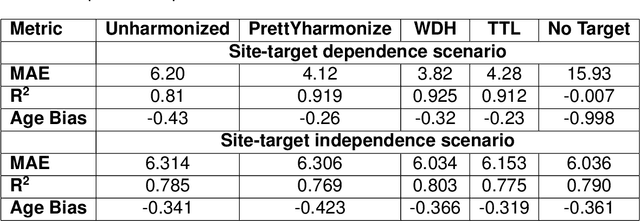
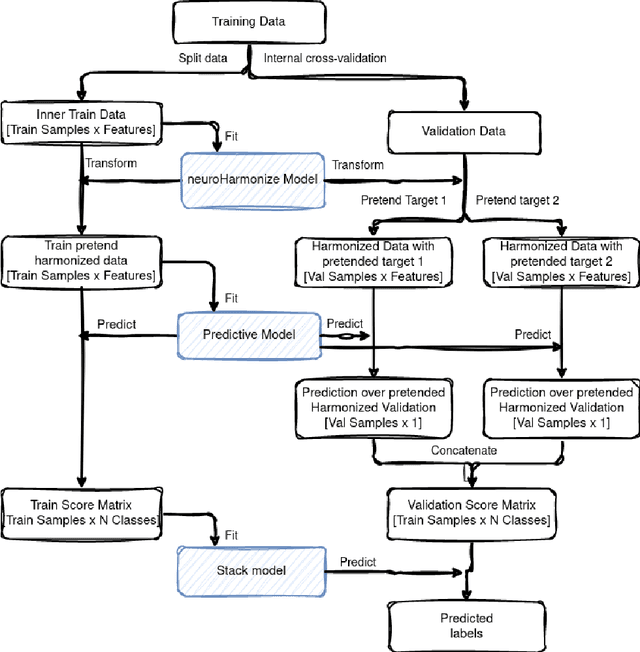
Abstract:Machine learning (ML) models benefit from large datasets. Collecting data in biomedical domains is costly and challenging, hence, combining datasets has become a common practice. However, datasets obtained under different conditions could present undesired site-specific variability. Data harmonization methods aim to remove site-specific variance while retaining biologically relevant information. This study evaluates the effectiveness of popularly used ComBat-based methods for harmonizing data in scenarios where the class balance is not equal across sites. We find that these methods struggle with data leakage issues. To overcome this problem, we propose a novel approach PrettYharmonize, designed to harmonize data by pretending the target labels. We validate our approach using controlled datasets designed to benchmark the utility of harmonization. Finally, using real-world MRI and clinical data, we compare leakage-prone methods with PrettYharmonize and show that it achieves comparable performance while avoiding data leakage, particularly in site-target-dependence scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge