Andrey Rzhetsky
Soft Sensing Transformer: Hundreds of Sensors are Worth a Single Word
Nov 10, 2021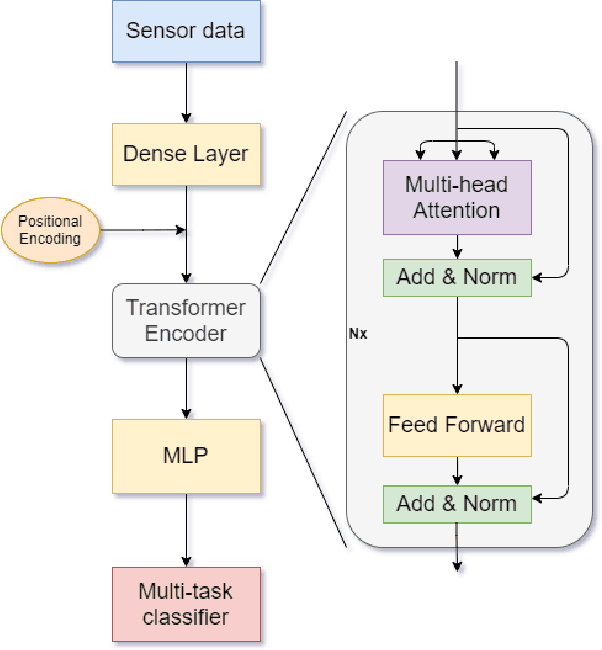
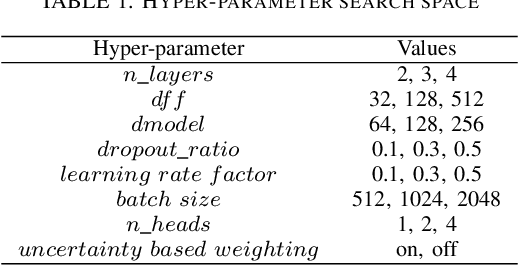
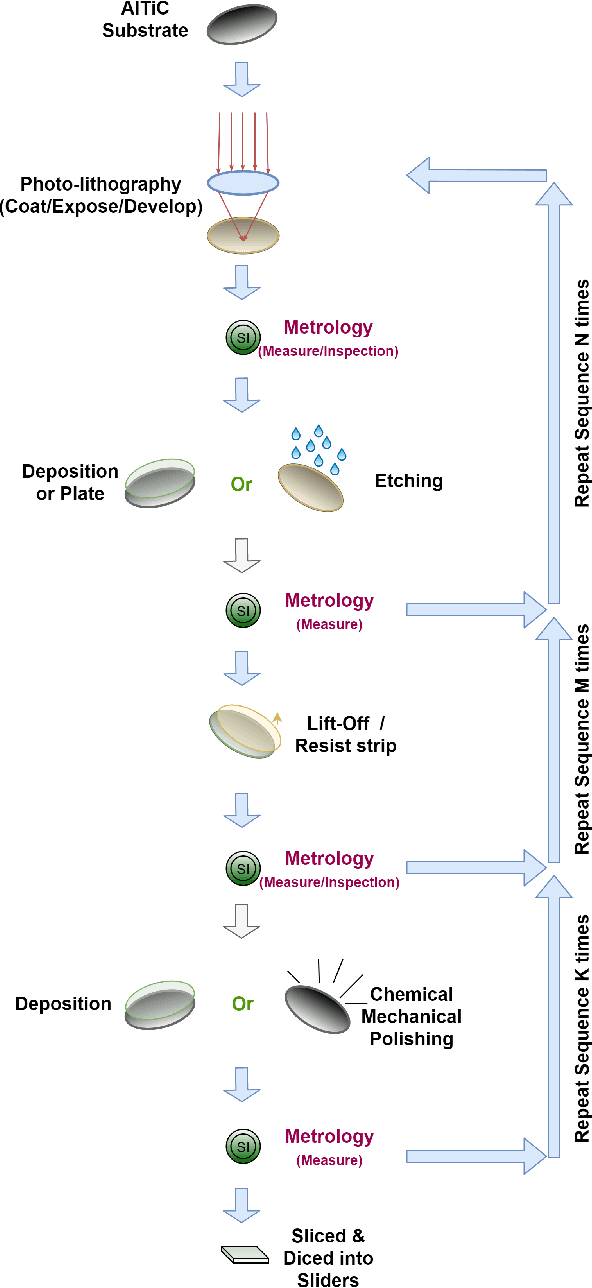
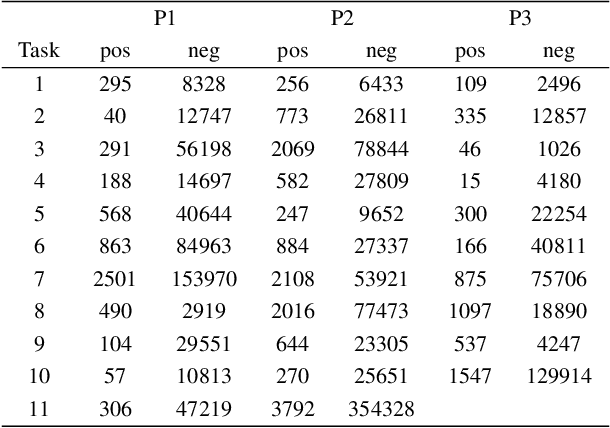
Abstract:With the rapid development of AI technology in recent years, there have been many studies with deep learning models in soft sensing area. However, the models have become more complex, yet, the data sets remain limited: researchers are fitting million-parameter models with hundreds of data samples, which is insufficient to exercise the effectiveness of their models and thus often fail to perform when implemented in industrial applications. To solve this long-lasting problem, we are providing large scale, high dimensional time series manufacturing sensor data from Seagate Technology to the public. We demonstrate the challenges and effectiveness of modeling industrial big data by a Soft Sensing Transformer model on these data sets. Transformer is used because, it has outperformed state-of-the-art techniques in Natural Language Processing, and since then has also performed well in the direct application to computer vision without introduction of image-specific inductive biases. We observe the similarity of a sentence structure to the sensor readings and process the multi-variable sensor readings in a time series in a similar manner of sentences in natural language. The high-dimensional time-series data is formatted into the same shape of embedded sentences and fed into the transformer model. The results show that transformer model outperforms the benchmark models in soft sensing field based on auto-encoder and long short-term memory (LSTM) models. To the best of our knowledge, we are the first team in academia or industry to benchmark the performance of original transformer model with large-scale numerical soft sensing data.
Lightweight Mobile Automated Assistant-to-physician for Global Lower-resource Areas
Oct 28, 2021Abstract:Importance: Lower-resource areas in Africa and Asia face a unique set of healthcare challenges: the dual high burden of communicable and non-communicable diseases; a paucity of highly trained primary healthcare providers in both rural and densely populated urban areas; and a lack of reliable, inexpensive internet connections. Objective: To address these challenges, we designed an artificial intelligence assistant to help primary healthcare providers in lower-resource areas document demographic and medical sign/symptom data and to record and share diagnostic data in real-time with a centralized database. Design: We trained our system using multiple data sets, including US-based electronic medical records (EMRs) and open-source medical literature and developed an adaptive, general medical assistant system based on machine learning algorithms. Main outcomes and Measure: The application collects basic information from patients and provides primary care providers with diagnoses and prescriptions suggestions. The application is unique from existing systems in that it covers a wide range of common diseases, signs, and medication typical in lower-resource countries; the application works with or without an active internet connection. Results: We have built and implemented an adaptive learning system that assists trained primary care professionals by means of an Android smartphone application, which interacts with a central database and collects real-time data. The application has been tested by dozens of primary care providers. Conclusions and Relevance: Our application would provide primary healthcare providers in lower-resource areas with a tool that enables faster and more accurate documentation of medical encounters. This application could be leveraged to automatically populate local or national EMR systems.
RIDDLE: Race and ethnicity Imputation from Disease history with Deep LEarning
Apr 27, 2018
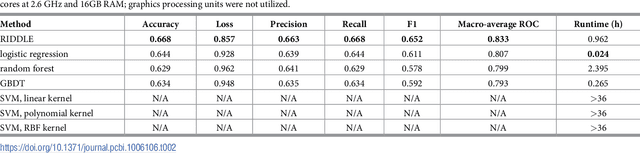

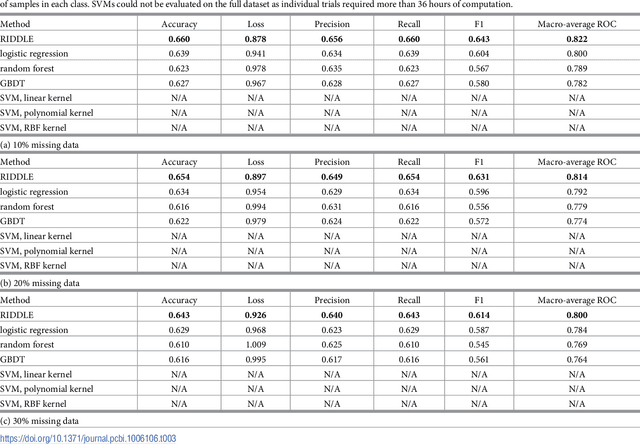
Abstract:Anonymized electronic medical records are an increasingly popular source of research data. However, these datasets often lack race and ethnicity information. This creates problems for researchers modeling human disease, as race and ethnicity are powerful confounders for many health exposures and treatment outcomes; race and ethnicity are closely linked to population-specific genetic variation. We showed that deep neural networks generate more accurate estimates for missing racial and ethnic information than competing methods (e.g., logistic regression, random forest). RIDDLE yielded significantly better classification performance across all metrics that were considered: accuracy, cross-entropy loss (error), and area under the curve for receiver operating characteristic plots (all $p < 10^{-6}$). We made specific efforts to interpret the trained neural network models to identify, quantify, and visualize medical features which are predictive of race and ethnicity. We used these characterizations of informative features to perform a systematic comparison of differential disease patterns by race and ethnicity. The fact that clinical histories are informative for imputing race and ethnicity could reflect (1) a skewed distribution of blue- and white-collar professions across racial and ethnic groups, (2) uneven accessibility and subjective importance of prophylactic health, (3) possible variation in lifestyle, such as dietary habits, and (4) differences in background genetic variation which predispose to diseases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge