Andreas Husch
Few-Shot Fingerprinting Subject Re-Identification in 3D-MRI and 2D-X-Ray
Dec 18, 2025Abstract:Combining open-source datasets can introduce data leakage if the same subject appears in multiple sets, leading to inflated model performance. To address this, we explore subject fingerprinting, mapping all images of a subject to a distinct region in latent space, to enable subject re-identification via similarity matching. Using a ResNet-50 trained with triplet margin loss, we evaluate few-shot fingerprinting on 3D MRI and 2D X-ray data in both standard (20-way 1-shot) and challenging (1000-way 1-shot) scenarios. The model achieves high Mean- Recall-@-K scores: 99.10% (20-way 1-shot) and 90.06% (500-way 5-shot) on ChestXray-14; 99.20% (20-way 1-shot) and 98.86% (100-way 3-shot) on BraTS- 2021.
DBSegment: Fast and robust segmentation of deep brain structures -- Evaluation of transportability across acquisition domains
Oct 18, 2021



Abstract:Segmenting deep brain structures from magnetic resonance images is important for patient diagnosis, surgical planning, and research. Most current state-of-the-art solutions follow a segmentation-by-registration approach, where subject MRIs are mapped to a template with well-defined segmentations. However, registration-based pipelines are time-consuming, thus, limiting their clinical use. This paper uses deep learning to provide a robust and efficient deep brain segmentation solution. The method consists of a pre-processing step to conform all MRI images to the same orientation, followed by a convolutional neural network using the nnU-Net framework. We use a total of 14 datasets from both research and clinical collections. Of these, seven were used for training and validation and seven were retained for independent testing. We trained the network to segment 30 deep brain structures, as well as a brain mask, using labels generated from a registration-based approach. We evaluated the generalizability of the network by performing a leave-one-dataset-out cross-validation, and extensive testing on external datasets. Furthermore, we assessed cross-domain transportability by evaluating the results separately on different domains. We achieved an average DSC of 0.89 $\pm$ 0.04 on the independent testing datasets when compared to the registration-based gold standard. On our test system, the computation time decreased from 42 minutes for a reference registration-based pipeline to 1 minute. Our proposed method is fast, robust, and generalizes with high reliability. It can be extended to the segmentation of other brain structures. The method is publicly available on GitHub, as well as a pip package for convenient usage.
A Solution for Multi-Alignment by Transformation Synchronisation
Apr 14, 2015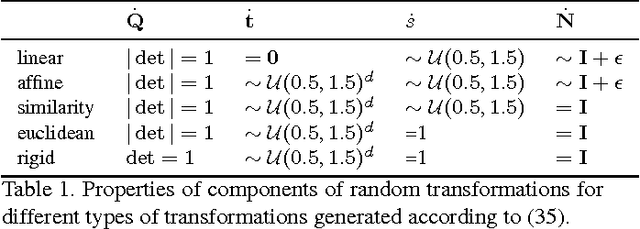
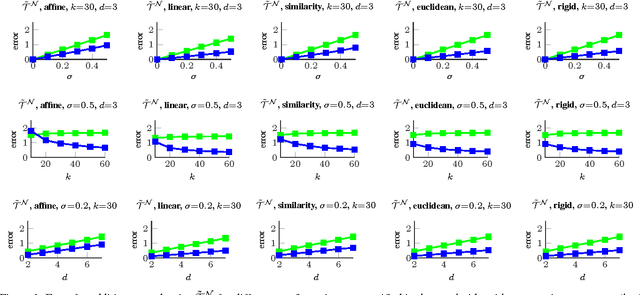


Abstract:The alignment of a set of objects by means of transformations plays an important role in computer vision. Whilst the case for only two objects can be solved globally, when multiple objects are considered usually iterative methods are used. In practice the iterative methods perform well if the relative transformations between any pair of objects are free of noise. However, if only noisy relative transformations are available (e.g. due to missing data or wrong correspondences) the iterative methods may fail. Based on the observation that the underlying noise-free transformations can be retrieved from the null space of a matrix that can directly be obtained from pairwise alignments, this paper presents a novel method for the synchronisation of pairwise transformations such that they are transitively consistent. Simulations demonstrate that for noisy transformations, a large proportion of missing data and even for wrong correspondence assignments the method delivers encouraging results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge