Amir Akbarnejad
MixFlow: Mixture-Conditioned Flow Matching for Out-of-Distribution Generalization
Jan 16, 2026Abstract:Achieving robust generalization under distribution shift remains a central challenge in conditional generative modeling, as existing conditional flow-based methods often struggle to extrapolate beyond the training conditions. We introduce MixFlow, a conditional flow-matching framework for descriptor-controlled generation that directly targets this limitation by jointly learning a descriptor-conditioned base distribution and a descriptor-conditioned flow field via shortest-path flow matching. By modeling the base distribution as a learnable, descriptor-dependent mixture, MixFlow enables smooth interpolation and extrapolation to unseen conditions, leading to substantially improved out-of-distribution generalization. We provide analytical insights into the behavior of the proposed framework and empirically demonstrate its effectiveness across multiple domains, including prediction of responses to unseen perturbations in single-cell transcriptomic data and high-content microscopy-based drug screening tasks. Across these diverse settings, MixFlow consistently outperforms standard conditional flow-matching baselines. Overall, MixFlow offers a simple yet powerful approach for achieving robust, generalizable, and controllable generative modeling across heterogeneous domains.
Predicting Ki67, ER, PR, and HER2 Statuses from H&E-stained Breast Cancer Images
Aug 03, 2023



Abstract:Despite the advances in machine learning and digital pathology, it is not yet clear if machine learning methods can accurately predict molecular information merely from histomorphology. In a quest to answer this question, we built a large-scale dataset (185538 images) with reliable measurements for Ki67, ER, PR, and HER2 statuses. The dataset is composed of mirrored images of H\&E and corresponding images of immunohistochemistry (IHC) assays (Ki67, ER, PR, and HER2. These images are mirrored through registration. To increase reliability, individual pairs were inspected and discarded if artifacts were present (tissue folding, bubbles, etc). Measurements for Ki67, ER and PR were determined by calculating H-Score from image analysis. HER2 measurement is based on binary classification: 0 and 1+ (IHC scores representing a negative subset) vs 3+ (IHC score positive subset). Cases with IHC equivocal score (2+) were excluded. We show that a standard ViT-based pipeline can achieve prediction performances around 90% in terms of Area Under the Curve (AUC) when trained with a proper labeling protocol. Finally, we shed light on the ability of the trained classifiers to localize relevant regions, which encourages future work to improve the localizations. Our proposed dataset is publicly available: https://ihc4bc.github.io/
GPEX, A Framework For Interpreting Artificial Neural Networks
Dec 18, 2021
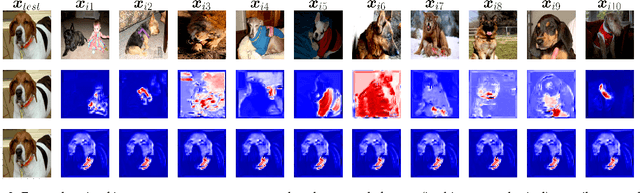
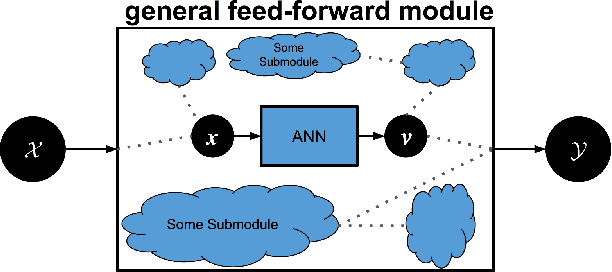
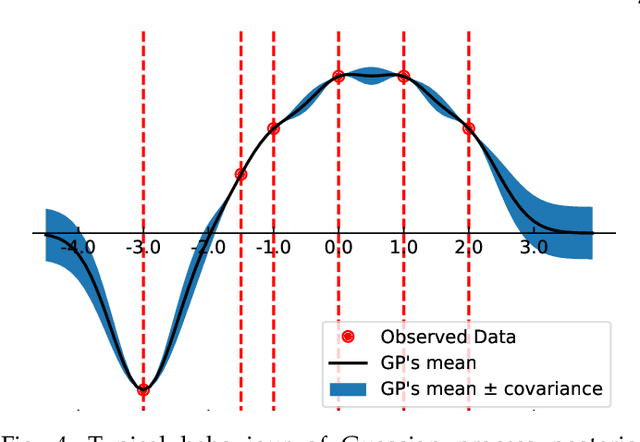
Abstract:Machine learning researchers have long noted a trade-off between interpretability and prediction performance. On the one hand, traditional models are often interpretable to humans but they cannot achieve high prediction performances. At the opposite end of the spectrum, deep models can achieve state-of-the-art performances in many tasks. However, deep models' predictions are known to be uninterpretable to humans. In this paper we present a framework that shortens the gap between the two aforementioned groups of methods. Given an artificial neural network (ANN), our method finds a Gaussian process (GP) whose predictions almost match those of the ANN. As GPs are highly interpretable, we use the trained GP to explain the ANN's decisions. We use our method to explain ANNs' decisions on may datasets. The explanations provide intriguing insights about the ANNs' decisions. With the best of our knowledge, our inference formulation for GPs is the first one in which an ANN and a similarly behaving Gaussian process naturally appear. Furthermore, we examine some of the known theoretical conditions under which an ANN is interpretable by GPs. Some of those theoretical conditions are too restrictive for modern architectures. However, we hypothesize that only a subset of those theoretical conditions are sufficient. Finally, we implement our framework as a publicly available tool called GPEX. Given any pytorch feed-forward module, GPEX allows users to interpret any ANN subcomponent of the module effortlessly and without having to be involved in the inference algorithm. GPEX is publicly available online:www.github.com/Nilanjan-Ray/gpex
Adversarial Attacks on Neural Networks for Graph Data
Jun 12, 2018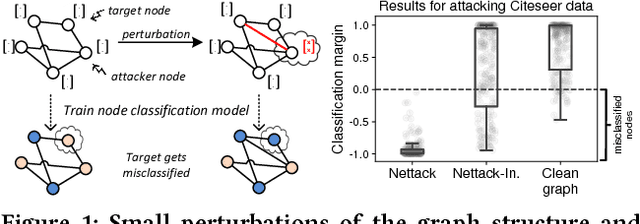
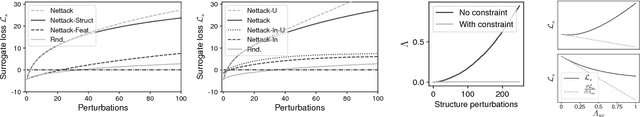

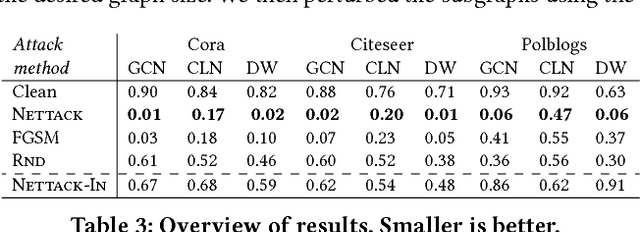
Abstract:Deep learning models for graphs have achieved strong performance for the task of node classification. Despite their proliferation, currently there is no study of their robustness to adversarial attacks. Yet, in domains where they are likely to be used, e.g. the web, adversaries are common. Can deep learning models for graphs be easily fooled? In this work, we introduce the first study of adversarial attacks on attributed graphs, specifically focusing on models exploiting ideas of graph convolutions. In addition to attacks at test time, we tackle the more challenging class of poisoning/causative attacks, which focus on the training phase of a machine learning model. We generate adversarial perturbations targeting the node's features and the graph structure, thus, taking the dependencies between instances in account. Moreover, we ensure that the perturbations remain unnoticeable by preserving important data characteristics. To cope with the underlying discrete domain we propose an efficient algorithm Nettack exploiting incremental computations. Our experimental study shows that accuracy of node classification significantly drops even when performing only few perturbations. Even more, our attacks are transferable: the learned attacks generalize to other state-of-the-art node classification models and unsupervised approaches, and likewise are successful even when only limited knowledge about the graph is given.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge