Aiham Taleb
Interpretable and Interactive Deep Multiple Instance Learning for Dental Caries Classification in Bitewing X-rays
Dec 17, 2021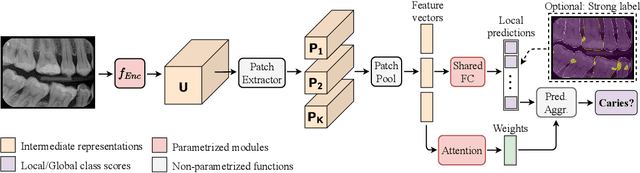
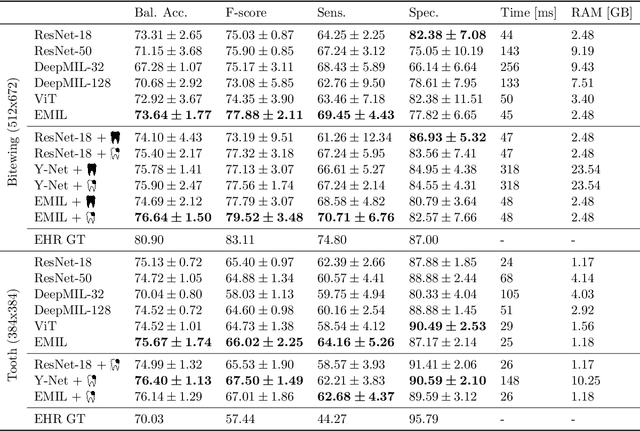
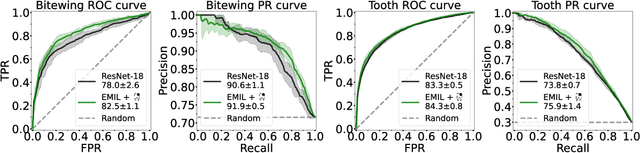
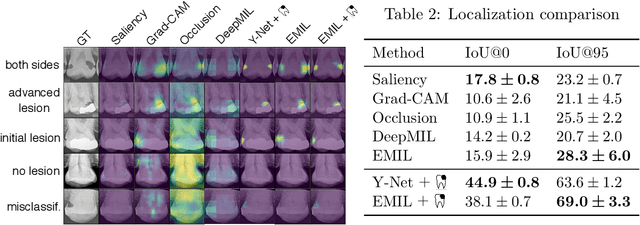
Abstract:We propose a simple and efficient image classification architecture based on deep multiple instance learning, and apply it to the challenging task of caries detection in dental radiographs. Technically, our approach contributes in two ways: First, it outputs a heatmap of local patch classification probabilities despite being trained with weak image-level labels. Second, it is amenable to learning from segmentation labels to guide training. In contrast to existing methods, the human user can faithfully interpret predictions and interact with the model to decide which regions to attend to. Experiments are conducted on a large clinical dataset of $\sim$38k bitewings ($\sim$316k teeth), where we achieve competitive performance compared to various baselines. When guided by an external caries segmentation model, a significant improvement in classification and localization performance is observed.
ContIG: Self-supervised Multimodal Contrastive Learning for Medical Imaging with Genetics
Nov 26, 2021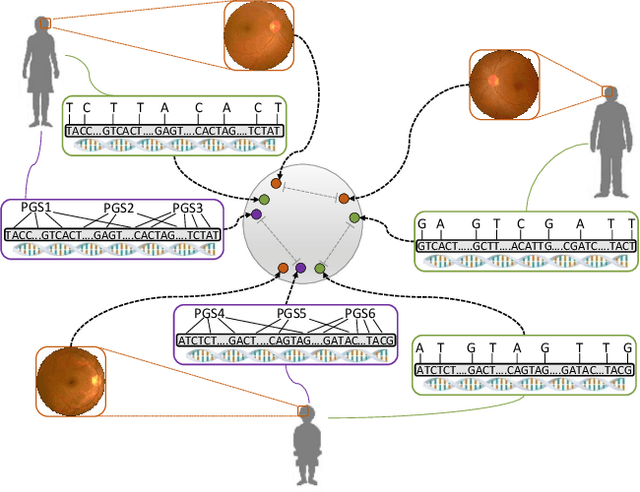
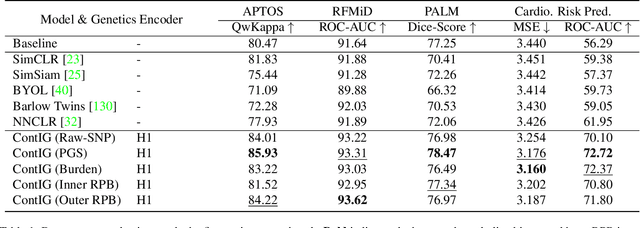
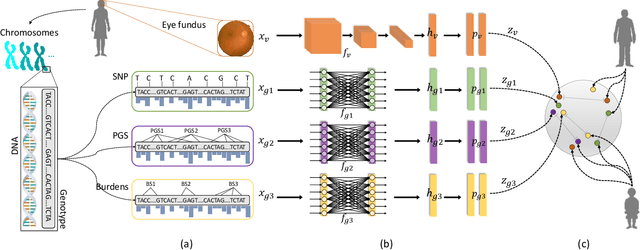
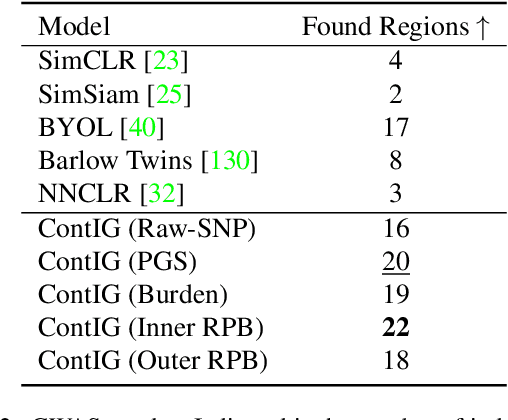
Abstract:High annotation costs are a substantial bottleneck in applying modern deep learning architectures to clinically relevant medical use cases, substantiating the need for novel algorithms to learn from unlabeled data. In this work, we propose ContIG, a self-supervised method that can learn from large datasets of unlabeled medical images and genetic data. Our approach aligns images and several genetic modalities in the feature space using a contrastive loss. We design our method to integrate multiple modalities of each individual person in the same model end-to-end, even when the available modalities vary across individuals. Our procedure outperforms state-of-the-art self-supervised methods on all evaluated downstream benchmark tasks. We also adapt gradient-based explainability algorithms to better understand the learned cross-modal associations between the images and genetic modalities. Finally, we perform genome-wide association studies on the features learned by our models, uncovering interesting relationships between images and genetic data.
Self-Supervised Learning for 3D Medical Image Analysis using 3D SimCLR and Monte Carlo Dropout
Oct 01, 2021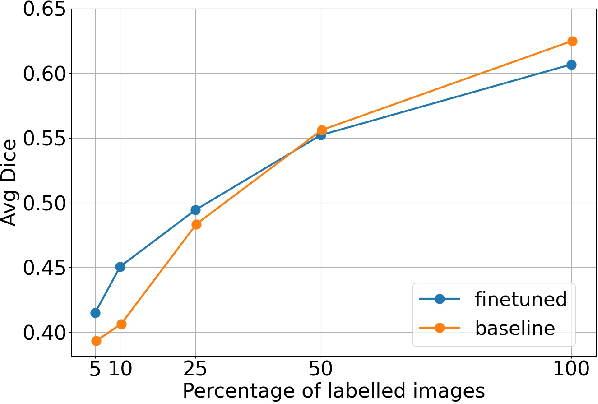
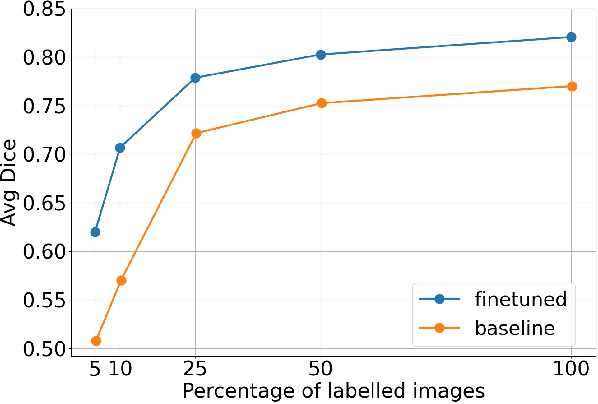
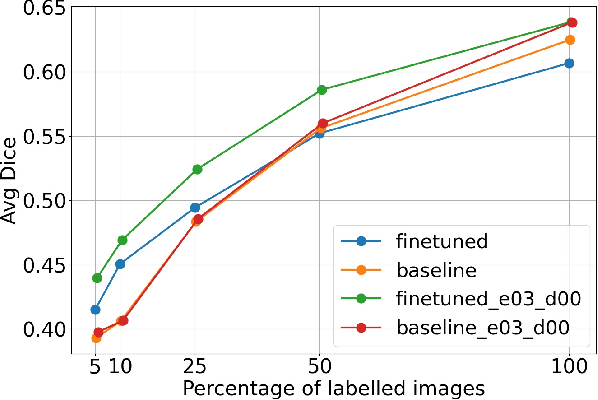
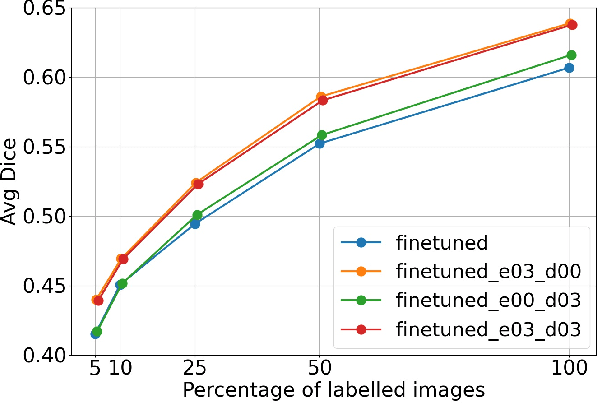
Abstract:Self-supervised learning methods can be used to learn meaningful representations from unlabeled data that can be transferred to supervised downstream tasks to reduce the need for labeled data. In this paper, we propose a 3D self-supervised method that is based on the contrastive (SimCLR) method. Additionally, we show that employing Bayesian neural networks (with Monte-Carlo Dropout) during the inference phase can further enhance the results on the downstream tasks. We showcase our models on two medical imaging segmentation tasks: i) Brain Tumor Segmentation from 3D MRI, ii) Pancreas Tumor Segmentation from 3D CT. Our experimental results demonstrate the benefits of our proposed methods in both downstream data-efficiency and performance.
3D Self-Supervised Methods for Medical Imaging
Jun 06, 2020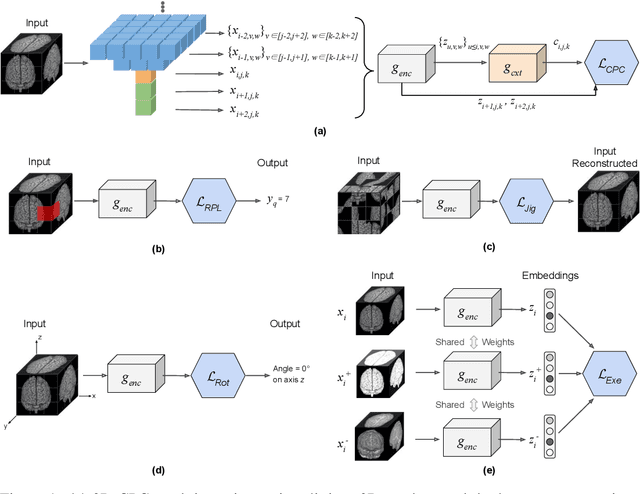
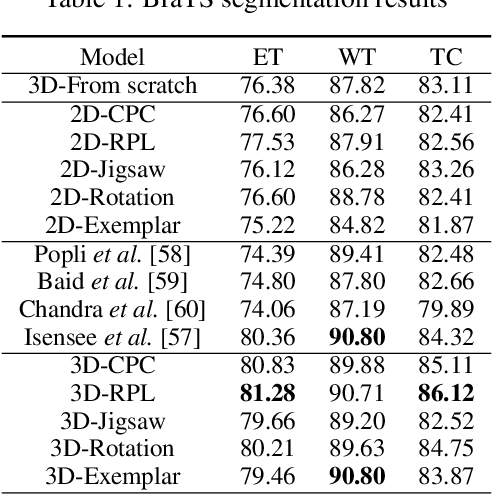
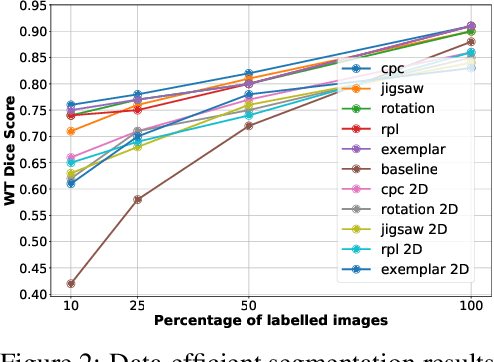

Abstract:Self-supervised learning methods have witnessed a recent surge of interest after proving successful in multiple application fields. In this work, we leverage these techniques, and we propose 3D versions for five different self-supervised methods, in the form of proxy tasks. Our methods facilitate neural network feature learning from unlabeled 3D images, aiming to reduce the required cost for expert annotation. The developed algorithms are 3D Contrastive Predictive Coding, 3D Rotation prediction, 3D Jigsaw puzzles, Relative 3D patch location, and 3D Exemplar networks. Our experiments show that pretraining models with our 3D tasks yields more powerful semantic representations, and enables solving downstream tasks more accurately and efficiently, compared to training the models from scratch and to pretraining them on 2D slices. We demonstrate the effectiveness of our methods on three downstream tasks from the medical imaging domain: i) Brain Tumor Segmentation from 3D MRI, ii) Pancreas Tumor Segmentation from 3D CT, and iii) Diabetic Retinopathy Detection from 2D Fundus images. In each task, we assess the gains in data-efficiency, performance, and speed of convergence. We achieve results competitive to state-of-the-art solutions at a fraction of the computational expense. We also publish the implementations for the 3D and 2D versions of our algorithms as an open-source library, in an effort to allow other researchers to apply and extend our methods on their datasets.
Multimodal Self-Supervised Learning for Medical Image Analysis
Dec 11, 2019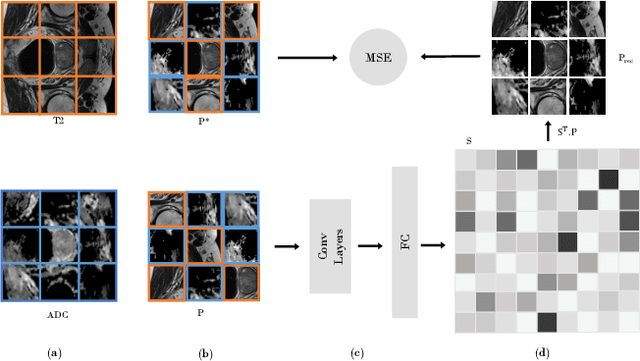
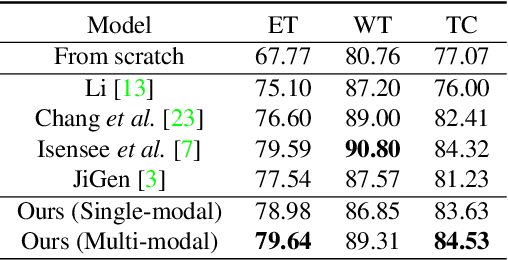
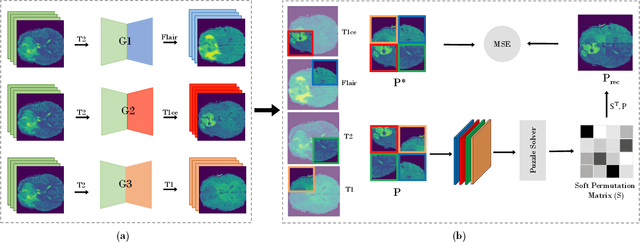
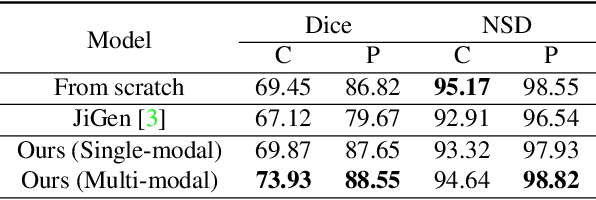
Abstract:In this paper, we propose a self-supervised learning approach that leverages multiple imaging modalities to increase data efficiency for medical image analysis. To this end, we introduce multimodal puzzle-solving proxy tasks, which facilitate neural network representation learning from multiple image modalities. These representations allow for subsequent fine-tuning on different downstream tasks. To achieve that, we employ the Sinkhorn operator to predict permutations of puzzle pieces in conjunction with a modality agnostic feature embedding. Together, they allow for a lean network architecture and increased computational efficiency. Under this framework, we propose different strategies for puzzle construction, integrating multiple medical imaging modalities, with varying levels of puzzle complexity. We benchmark these strategies in a range of experiments to assess the gains of our method in downstream performance and data-efficiency on different target tasks. Our experiments show that solving puzzles interleaved with multimodal content yields more powerful semantic representations. This allows us to solve downstream tasks more accurately and efficiently, compared to treating each modality independently. We demonstrate the effectiveness of the proposed approach on two multimodal medical imaging benchmarks: the BraTS and the Prostate semantic segmentation datasets, on which we achieve competitive results to state-of-the-art solutions, at a fraction of the computational expense. We also outperform many previous solutions on the chosen benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge