Ahmed Bilal Ashraf
COVIDomaly: A Deep Convolutional Autoencoder Approach for Detecting Early Cases of COVID-19
Oct 06, 2020



Abstract:As of September 2020, the COVID-19 pandemic continues to devastate the health and well-being of the global population. With more than 33 million confirmed cases and over a million deaths, global health organizations are still a long way from fully containing the pandemic. This pandemic has raised serious questions about the emergency preparedness of health agencies, not only in terms of treatment of an unseen disease, but also in identifying its early symptoms. In the particular case of COVID-19, several studies have indicated that chest radiography images of the infected patients show characteristic abnormalities. However, at the onset of a given pandemic, such as COVID-19, there may not be sufficient data for the affected cases to train models for their robust detection. Hence, supervised classification is ill-posed for this problem because the time spent in collecting large amounts of infected peoples' data could lead to the loss of human lives and delays in preventive interventions. Therefore, we formulate this problem within a one-class classification framework, in which the data for healthy patients is abundantly available, whereas no training data is present for the class of interest (COVID-19 in our case). To solve this problem, we present COVIDomaly, a convolutional autoencoder framework to detect unseen COVID-19 cases from the chest radiographs. We tested two settings on a publicly available dataset (COVIDx) by training the model on chest X-rays from (i) only healthy adults, and (ii) healthy and other non-COVID-19 pneumonia, and detected COVID-19 as an anomaly. After performing 3-fold cross validation, we obtain a pooled ROC-AUC of 0.7652 and 0.6902 in the two settings respectively. These results are very encouraging and pave the way towards research for ensuring emergency preparedness in future pandemics, especially the ones that could be detected from chest X-rays.
A Hybrid Instance-based Transfer Learning Method
Dec 03, 2018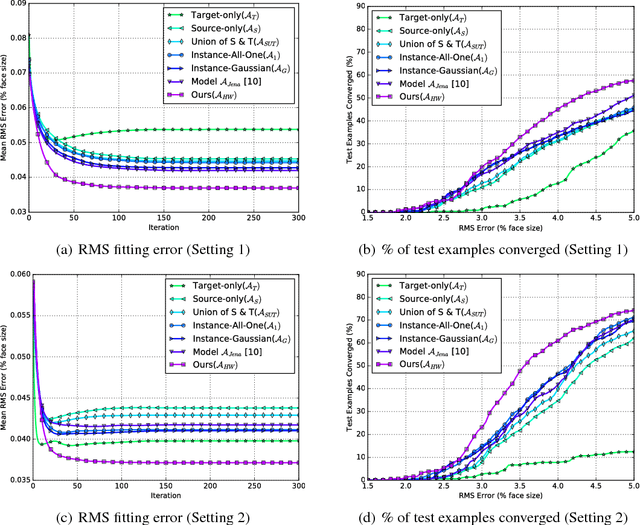
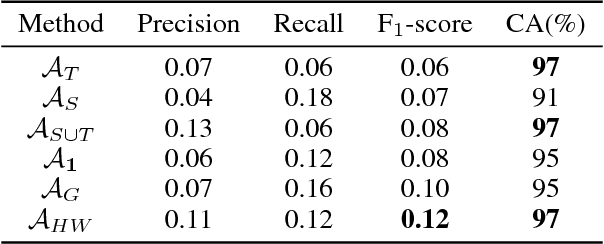
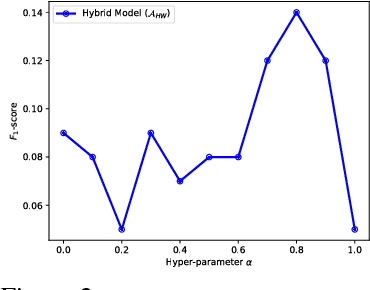

Abstract:In recent years, supervised machine learning models have demonstrated tremendous success in a variety of application domains. Despite the promising results, these successful models are data hungry and their performance relies heavily on the size of training data. However, in many healthcare applications it is difficult to collect sufficiently large training datasets. Transfer learning can help overcome this issue by transferring the knowledge from readily available datasets (source) to a new dataset (target). In this work, we propose a hybrid instance-based transfer learning method that outperforms a set of baselines including state-of-the-art instance-based transfer learning approaches. Our method uses a probabilistic weighting strategy to fuse information from the source domain to the model learned in the target domain. Our method is generic, applicable to multiple source domains, and robust with respect to negative transfer. We demonstrate the effectiveness of our approach through extensive experiments for two different applications.
Subspace Selection to Suppress Confounding Source Domain Information in AAM Transfer Learning
Oct 03, 2017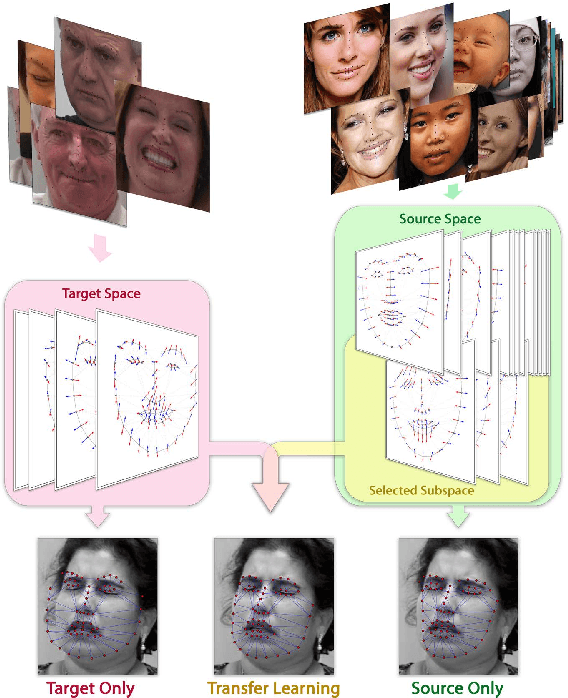
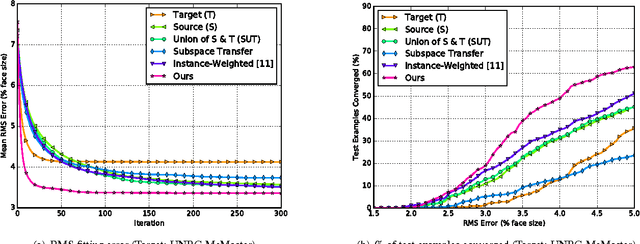
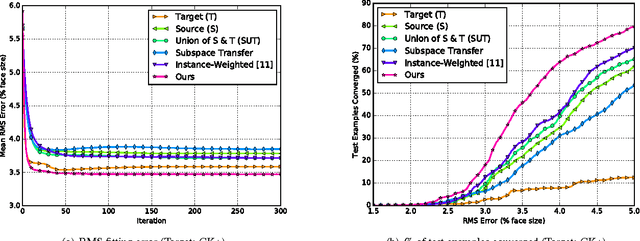
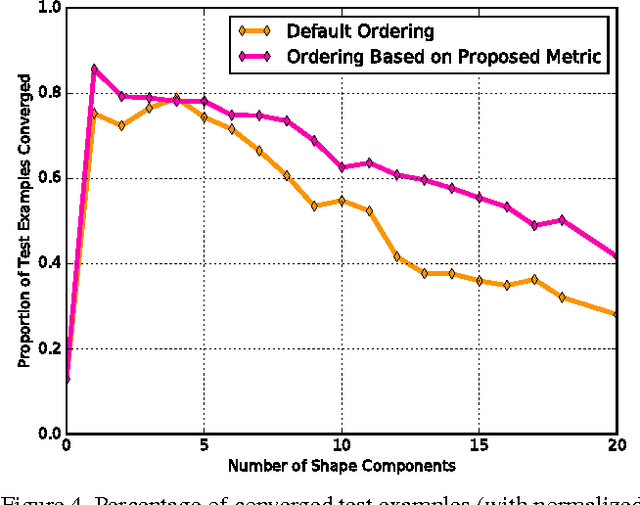
Abstract:Active appearance models (AAMs) are a class of generative models that have seen tremendous success in face analysis. However, model learning depends on the availability of detailed annotation of canonical landmark points. As a result, when accurate AAM fitting is required on a different set of variations (expression, pose, identity), a new dataset is collected and annotated. To overcome the need for time consuming data collection and annotation, transfer learning approaches have received recent attention. The goal is to transfer knowledge from previously available datasets (source) to a new dataset (target). We propose a subspace transfer learning method, in which we select a subspace from the source that best describes the target space. We propose a metric to compute the directional similarity between the source eigenvectors and the target subspace. We show an equivalence between this metric and the variance of target data when projected onto source eigenvectors. Using this equivalence, we select a subset of source principal directions that capture the variance in target data. To define our model, we augment the selected source subspace with the target subspace learned from a handful of target examples. In experiments done on six publicly available datasets, we show that our approach outperforms the state of the art in terms of the RMS fitting error as well as the percentage of test examples for which AAM fitting converges to the ground truth.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge