Abhay Shah
3-D Surface Segmentation Meets Conditional Random Fields
Jun 11, 2019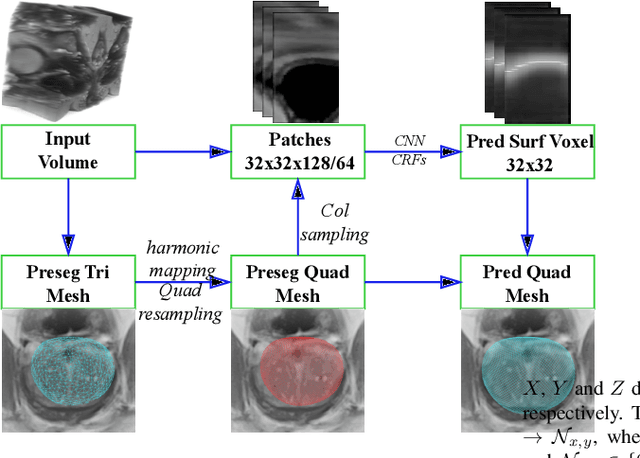
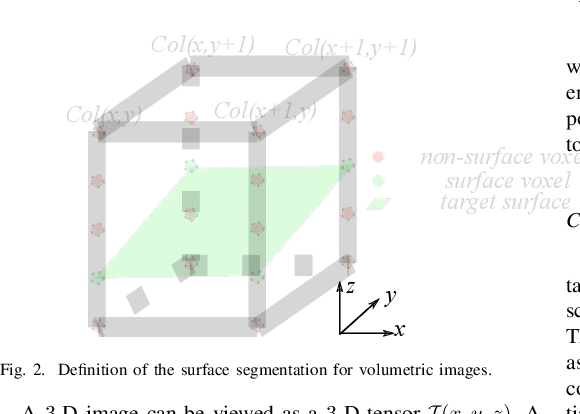
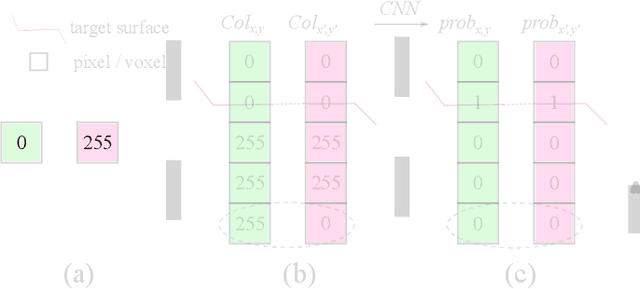
Abstract:Automated surface segmentation is important and challenging in many medical image analysis applications. Recent deep learning based methods have been developed for various object segmentation tasks. Most of them are a classification based approach, e.g. U-net, which predicts the probability of being target object or background for each voxel. One problem of those methods is lacking of topology guarantee for segmented objects, and usually post processing is needed to infer the boundary surface of the object. In this paper, a novel model based on 3-D convolutional neural network (CNN) and Conditional Random Fields (CRFs) is proposed to tackle the surface segmentation problem with end-to-end training. To the best of our knowledge, this is the first study to apply a 3-D neural network with a CRFs model for direct surface segmentation. Experiments carried out on NCI-ISBI 2013 MR prostate dataset and Medical Segmentation Decathlon Spleen dataset demonstrated very promising segmentation results.
Optimal Multi-Object Segmentation with Novel Gradient Vector Flow Based Shape Priors
May 22, 2017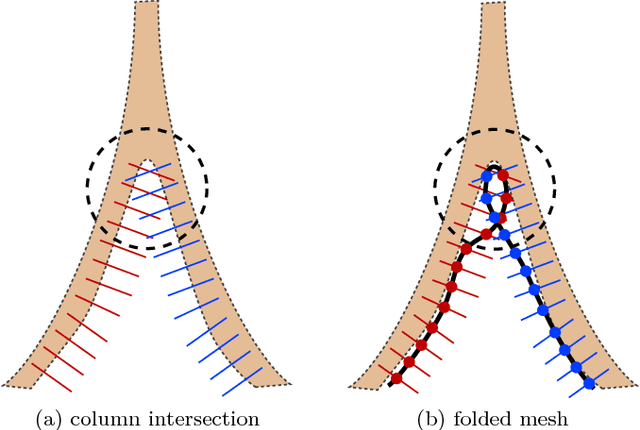
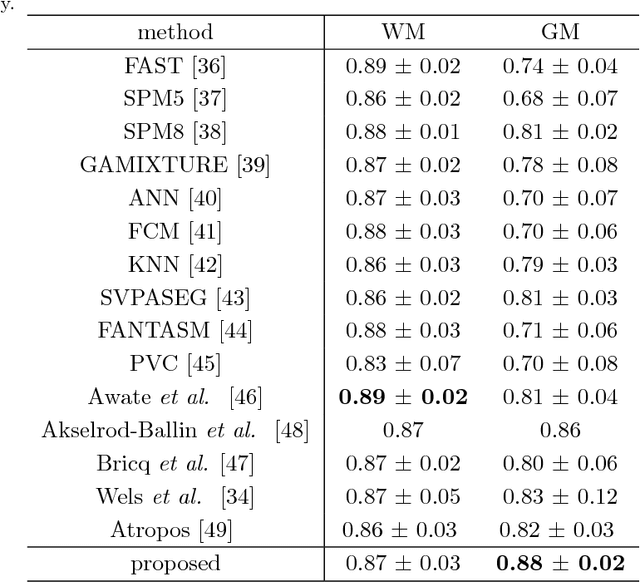
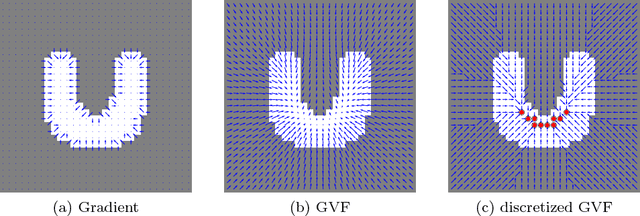
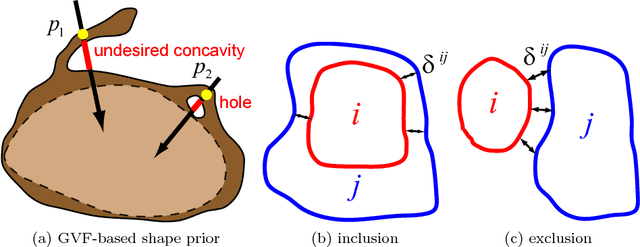
Abstract:Shape priors have been widely utilized in medical image segmentation to improve segmentation accuracy and robustness. A major way to encode such a prior shape model is to use a mesh representation, which is prone to causing self-intersection or mesh folding. Those problems require complex and expensive algorithms to mitigate. In this paper, we propose a novel shape prior directly embedded in the voxel grid space, based on gradient vector flows of a pre-segmentation. The flexible and powerful prior shape representation is ready to be extended to simultaneously segmenting multiple interacting objects with minimum separation distance constraint. The problem is formulated as a Markov random field problem whose exact solution can be efficiently computed with a single minimum s-t cut in an appropriately constructed graph. The proposed algorithm is validated on two multi-object segmentation applications: the brain tissue segmentation in MRI images, and the bladder/prostate segmentation in CT images. Both sets of experiments show superior or competitive performance of the proposed method to other state-of-the-art methods.
Simultaneous Multiple Surface Segmentation Using Deep Learning
May 19, 2017



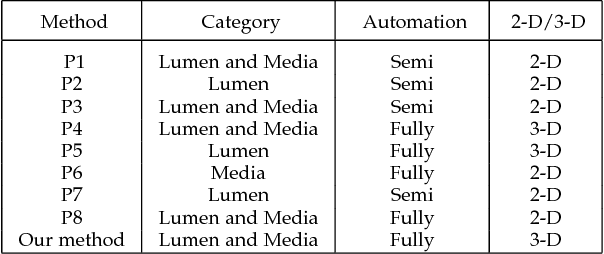
Abstract:The task of automatically segmenting 3-D surfaces representing boundaries of objects is important for quantitative analysis of volumetric images, and plays a vital role in biomedical image analysis. Recently, graph-based methods with a global optimization property have been developed and optimized for various medical imaging applications. Despite their widespread use, these require human experts to design transformations, image features, surface smoothness priors, and re-design for a different tissue, organ or imaging modality. Here, we propose a Deep Learning based approach for segmentation of the surfaces in volumetric medical images, by learning the essential features and transformations from training data, without any human expert intervention. We employ a regional approach to learn the local surface profiles. The proposed approach was evaluated on simultaneous intraretinal layer segmentation of optical coherence tomography (OCT) images of normal retinas and retinas affected by age related macular degeneration (AMD). The proposed approach was validated on 40 retina OCT volumes including 20 normal and 20 AMD subjects. The experiments showed statistically significant improvement in accuracy for our approach compared to state-of-the-art graph based optimal surface segmentation with convex priors (G-OSC). A single Convolution Neural Network (CNN) was used to learn the surfaces for both normal and diseased images. The mean unsigned surface positioning errors obtained by G-OSC method 2.31 voxels (95% CI 2.02-2.60 voxels) was improved to $1.27$ voxels (95% CI 1.14-1.40 voxels) using our new approach. On average, our approach takes 94.34 s, requiring 95.35 MB memory, which is much faster than the 2837.46 s and 6.87 GB memory required by the G-OSC method on the same computer system.
Optimal Multiple Surface Segmentation with Convex Priors in Irregularly Sampled Space
May 16, 2017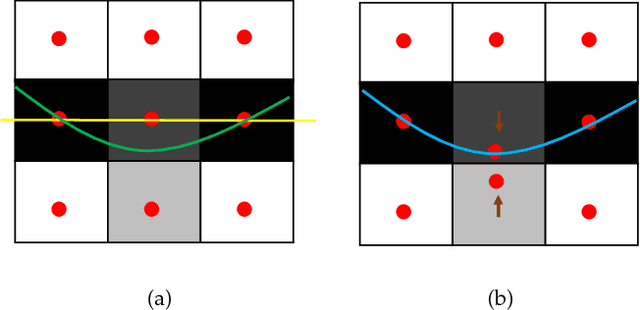
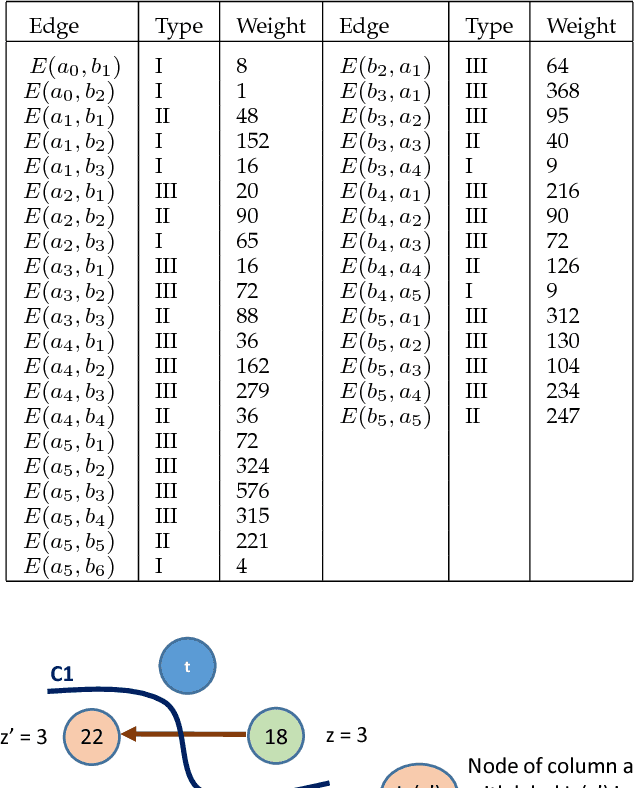
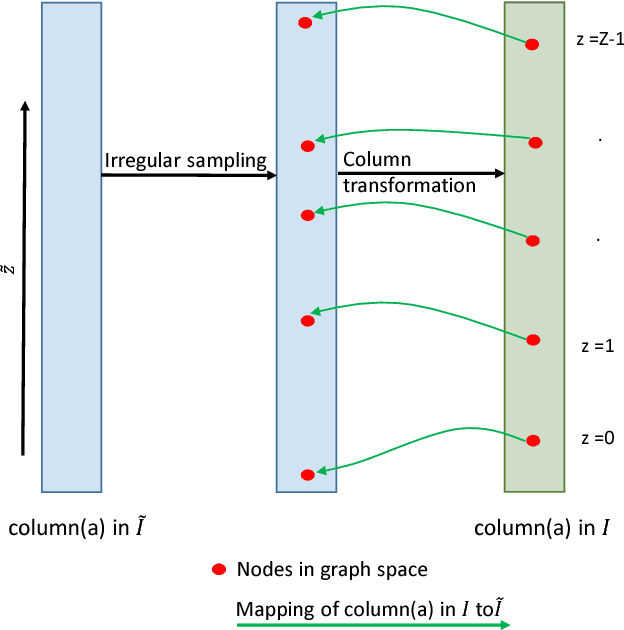
Abstract:Optimal surface segmentation is widely used in numerous medical image segmentation applications. However, nodes in the graph based optimal surface segmentation method typically encode uniformly distributed orthogonal voxels of the volume. Thus the segmentation cannot attain an accuracy greater than a single unit voxel, i.e. the distance between two adjoining nodes in graph space. Segmentation accuracy higher than a unit voxel is achievable by exploiting partial volume information in the voxels which shall result in non-equidistant spacing between adjoining graph nodes. This paper reports a generalized graph based optimal multiple surface segmentation method with convex priors which segments the target surfaces in irregularly sampled space. The proposed method allows non-equidistant spacing between the adjoining graph nodes to achieve subvoxel accurate segmentation by utilizing the partial volume information in the voxels. The partial volume information in the voxels is exploited by computing a displacement field from the original volume data to identify the subvoxel accurate centers within each voxel resulting in non-equidistant spacing between the adjoining graph nodes. The smoothness of each surface modelled as a convex constraint governs the connectivity and regularity of the surface. We employ an edge-based graph representation to incorporate the necessary constraints and the globally optimal solution is obtained by computing a minimum s-t cut. The proposed method was validated on 25 optical coherence tomography image volumes of the retina and 10 intravascular multi-frame ultrasound image datasets for subvoxel and super resolution segmentation accuracy. In all cases, the approach yielded highly accurate results. Our approach can be readily extended to higher-dimensional segmentations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge