XY Network for Nuclear Segmentation in Multi-Tissue Histology Images
Paper and Code
Dec 16, 2018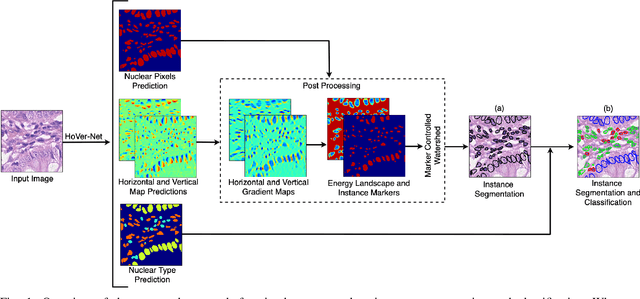
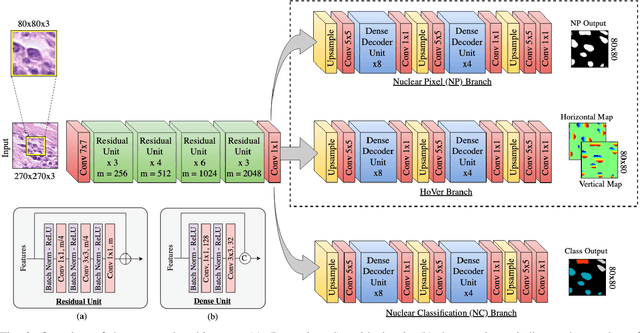
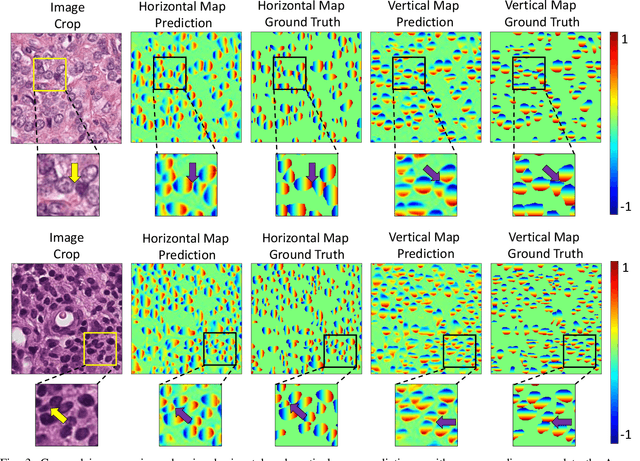
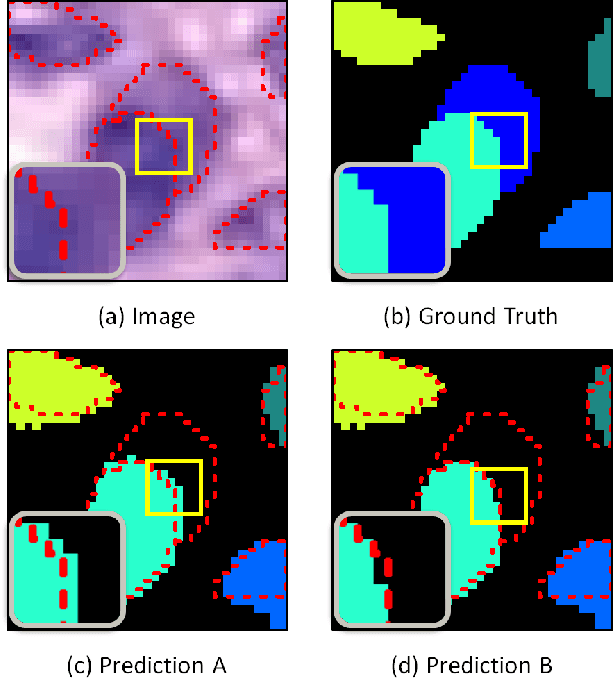
Nuclear segmentation within Haematoxylin & Eosin stained histology images is a fundamental prerequisite in the digital pathology work-flow, due to the ability for nuclear features to act as key diagnostic markers. The development of automated methods for nuclear segmentation enables the quantitative analysis of tens of thousands of nuclei within a whole-slide pathology image, opening up possibilities of further analysis of large-scale nuclear morphometry. However, automated nuclear segmentation is faced with a major challenge in that there are several different types of nuclei, some of them exhibiting large intra-class variability such as the tumour cells. Additionally, some of the nuclei are often clustered together. To address these challenges, we present a novel convolutional neural network for automated nuclear segmentation that leverages the instance-rich information encoded within the vertical and horizontal distances of nuclear pixels to their centres of mass. These distances are then utilised to separate clustered nuclei, resulting in an accurate segmentation, particularly in areas with overlapping instances. We demonstrate state-of-the-art performance compared to other methods on four independent multi-tissue histology image datasets. Furthermore, we propose an interpretable and reliable evaluation framework that effectively quantifies nuclear segmentation performance and overcomes the limitations of existing performance measures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge