Transfer Learning for Clinical Time Series Analysis using Deep Neural Networks
Paper and Code
Apr 01, 2019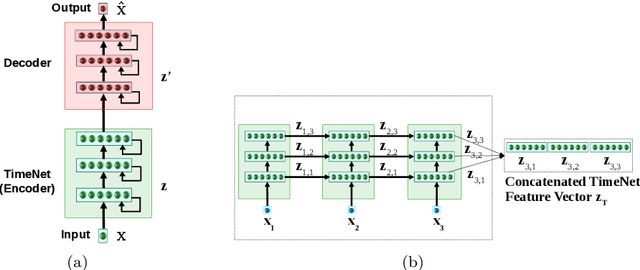
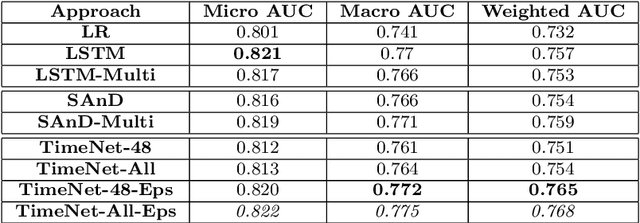
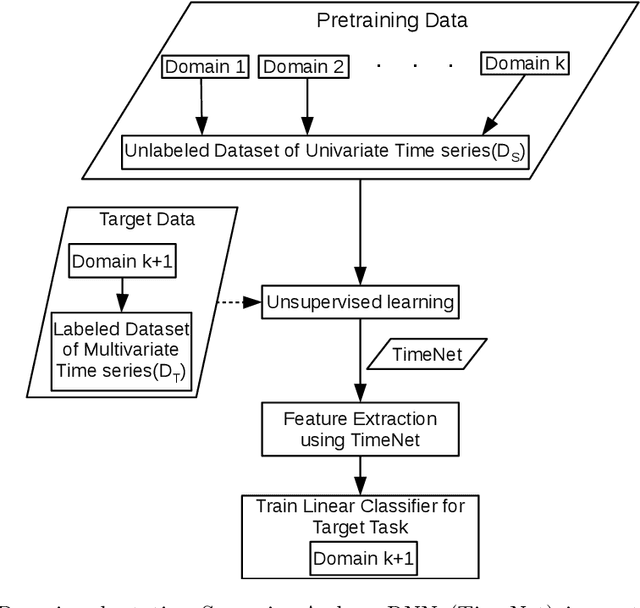

Deep neural networks have shown promising results for various clinical prediction tasks. However, training deep networks such as those based on Recurrent Neural Networks (RNNs) requires large labeled data, significant hyper-parameter tuning effort and expertise, and high computational resources. In this work, we investigate as to what extent can transfer learning address these issues when using deep RNNs to model multivariate clinical time series. We consider two scenarios for transfer learning using RNNs: i) domain-adaptation, i.e., leveraging a deep RNN - namely, TimeNet - pre-trained for feature extraction on time series from diverse domains, and adapting it for feature extraction and subsequent target tasks in healthcare domain, ii) task-adaptation, i.e., pre-training a deep RNN - namely, HealthNet - on diverse tasks in healthcare domain, and adapting it to new target tasks in the same domain. We evaluate the above approaches on publicly available MIMIC-III benchmark dataset, and demonstrate that (a) computationally-efficient linear models trained using features extracted via pre-trained RNNs outperform or, in the worst case, perform as well as deep RNNs and statistical hand-crafted features based models trained specifically for target task; (b) models obtained by adapting pre-trained models for target tasks are significantly more robust to the size of labeled data compared to task-specific RNNs, while also being computationally efficient. We, therefore, conclude that pre-trained deep models like TimeNet and HealthNet allow leveraging the advantages of deep learning for clinical time series analysis tasks, while also minimize dependence on hand-crafted features, deal robustly with scarce labeled training data scenarios without overfitting, as well as reduce dependence on expertise and resources required to train deep networks from scratch.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge