SIRE: scale-invariant, rotation-equivariant estimation of artery orientations using graph neural networks
Paper and Code
Nov 09, 2023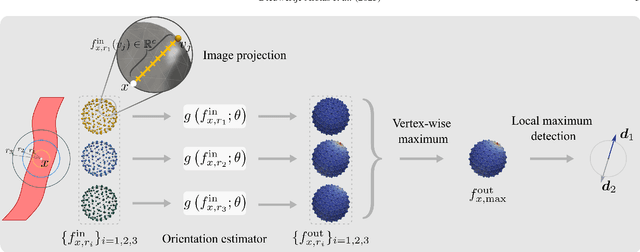
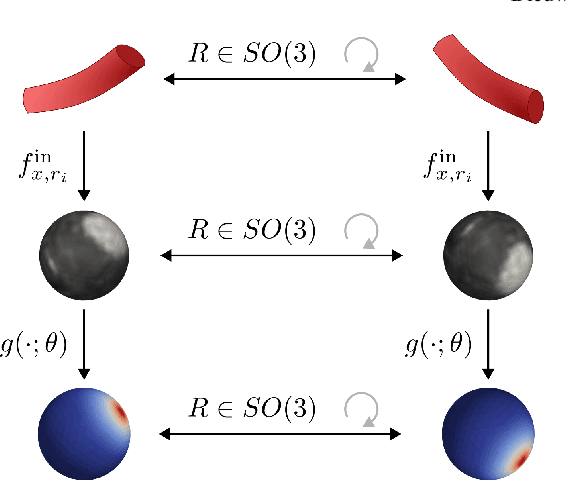
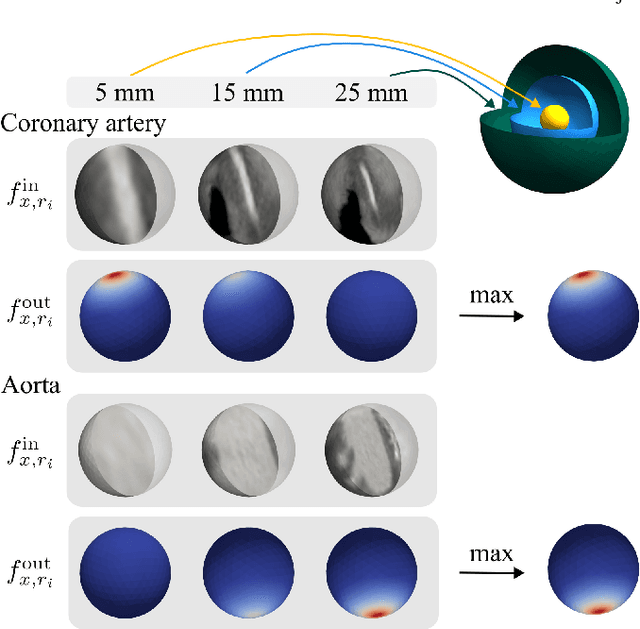
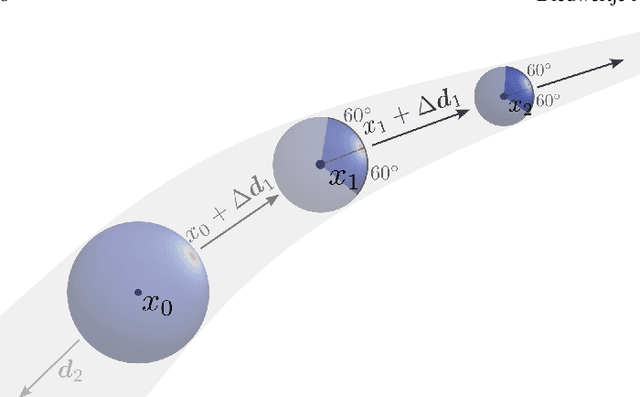
Blood vessel orientation as visualized in 3D medical images is an important descriptor of its geometry that can be used for centerline extraction and subsequent segmentation and visualization. Arteries appear at many scales and levels of tortuosity, and determining their exact orientation is challenging. Recent works have used 3D convolutional neural networks (CNNs) for this purpose, but CNNs are sensitive to varying vessel sizes and orientations. We present SIRE: a scale-invariant, rotation-equivariant estimator for local vessel orientation. SIRE is modular and can generalise due to symmetry preservation. SIRE consists of a gauge equivariant mesh CNN (GEM-CNN) operating on multiple nested spherical meshes with different sizes in parallel. The features on each mesh are a projection of image intensities within the corresponding sphere. These features are intrinsic to the sphere and, in combination with the GEM-CNN, lead to SO(3)-equivariance. Approximate scale invariance is achieved by weight sharing and use of a symmetric maximum function to combine multi-scale predictions. Hence, SIRE can be trained with arbitrarily oriented vessels with varying radii to generalise to vessels with a wide range of calibres and tortuosity. We demonstrate the efficacy of SIRE using three datasets containing vessels of varying scales: the vascular model repository (VMR), the ASOCA coronary artery set, and a set of abdominal aortic aneurysms (AAAs). We embed SIRE in a centerline tracker which accurately tracks AAAs, regardless of the data SIRE is trained with. Moreover, SIRE can be used to track coronary arteries, even when trained only with AAAs. In conclusion, by incorporating SO(3) and scale symmetries, SIRE can determine the orientations of vessels outside of the training domain, forming a robust and data-efficient solution to geometric analysis of blood vessels in 3D medical images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge