SiamQuality: A ConvNet-Based Foundation Model for Imperfect Physiological Signals
Paper and Code
Apr 26, 2024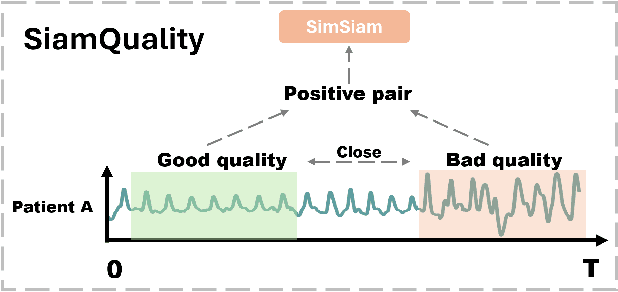

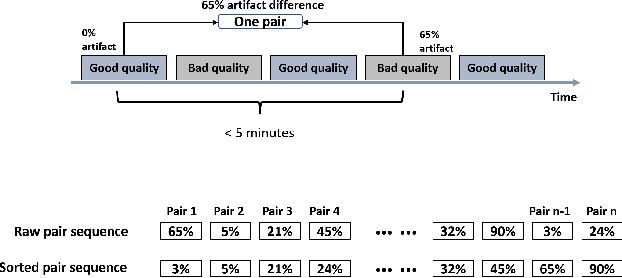
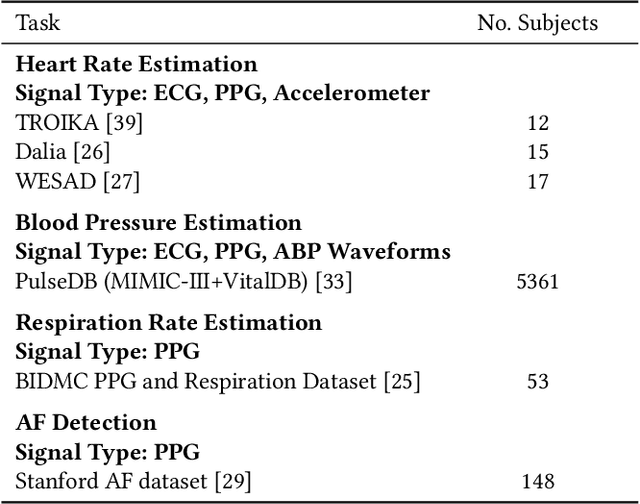
Foundation models, especially those using transformers as backbones, have gained significant popularity, particularly in language and language-vision tasks. However, large foundation models are typically trained on high-quality data, which poses a significant challenge, given the prevalence of poor-quality real-world data. This challenge is more pronounced for developing foundation models for physiological data; such data are often noisy, incomplete, or inconsistent. The present work aims to provide a toolset for developing foundation models on physiological data. We leverage a large dataset of photoplethysmography (PPG) signals from hospitalized intensive care patients. For this data, we propose SimQuality, a novel self-supervised learning task based on convolutional neural networks (CNNs) as the backbone to enforce representations to be similar for good and poor quality signals that are from similar physiological states. We pre-trained the SimQuality on over 36 million 30-second PPG pairs and then fine-tuned and tested on six downstream tasks using external datasets. The results demonstrate the superiority of the proposed approach on all the downstream tasks, which are extremely important for heart monitoring on wearable devices. Our method indicates that CNNs can be an effective backbone for foundation models that are robust to training data quality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge