Riemannian-geometry-based modeling and clustering of network-wide non-stationary time series: The brain-network case
Paper and Code
Jan 26, 2017
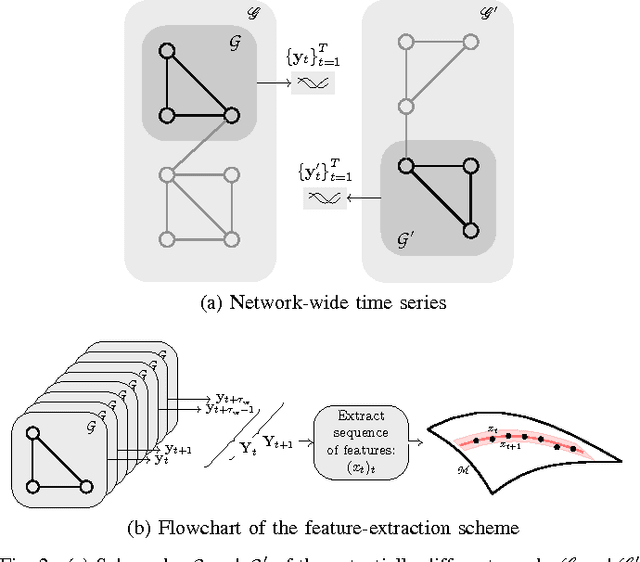
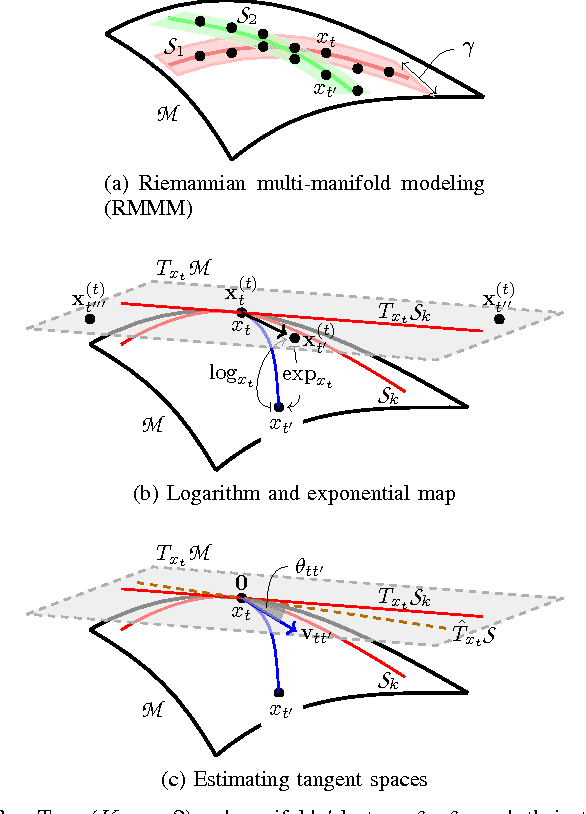
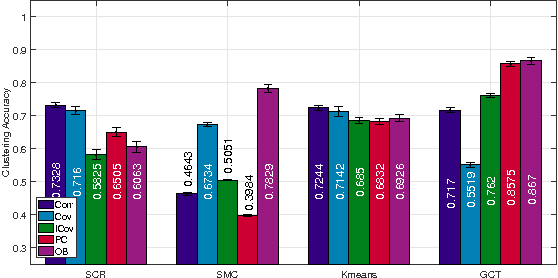
This paper advocates Riemannian multi-manifold modeling in the context of network-wide non-stationary time-series analysis. Time-series data, collected sequentially over time and across a network, yield features which are viewed as points in or close to a union of multiple submanifolds of a Riemannian manifold, and distinguishing disparate time series amounts to clustering multiple Riemannian submanifolds. To support the claim that exploiting the latent Riemannian geometry behind many statistical features of time series is beneficial to learning from network data, this paper focuses on brain networks and puts forth two feature-generation schemes for network-wide dynamic time series. The first is motivated by Granger-causality arguments and uses an auto-regressive moving average model to map low-rank linear vector subspaces, spanned by column vectors of appropriately defined observability matrices, to points into the Grassmann manifold. The second utilizes (non-linear) dependencies among network nodes by introducing kernel-based partial correlations to generate points in the manifold of positive-definite matrices. Capitilizing on recently developed research on clustering Riemannian submanifolds, an algorithm is provided for distinguishing time series based on their geometrical properties, revealed within Riemannian feature spaces. Extensive numerical tests demonstrate that the proposed framework outperforms classical and state-of-the-art techniques in clustering brain-network states/structures hidden beneath synthetic fMRI time series and brain-activity signals generated from real brain-network structural connectivity matrices.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge