Predicting single-cell perturbation responses for unseen drugs
Paper and Code
Apr 28, 2022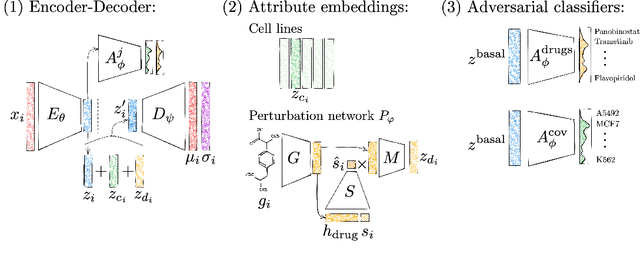
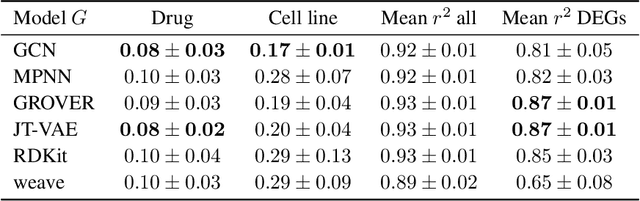
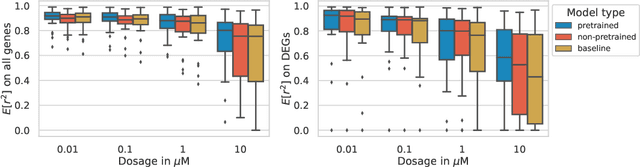
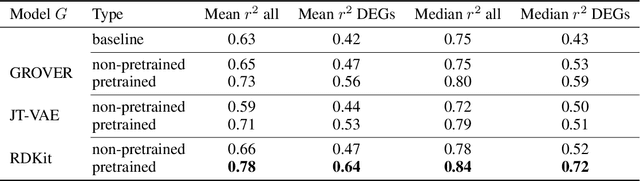
Single-cell transcriptomics enabled the study of cellular heterogeneity in response to perturbations at the resolution of individual cells. However, scaling high-throughput screens (HTSs) to measure cellular responses for many drugs remains a challenge due to technical limitations and, more importantly, the cost of such multiplexed experiments. Thus, transferring information from routinely performed bulk RNA-seq HTS is required to enrich single-cell data meaningfully. We introduce a new encoder-decoder architecture to study the perturbational effects of unseen drugs. We combine the model with a transfer learning scheme and demonstrate how training on existing bulk RNA-seq HTS datasets can improve generalisation performance. Better generalisation reduces the need for extensive and costly screens at single-cell resolution. We envision that our proposed method will facilitate more efficient experiment designs through its ability to generate in-silico hypotheses, ultimately accelerating targeted drug discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge