One Transformer Can Understand Both 2D & 3D Molecular Data
Paper and Code
Oct 04, 2022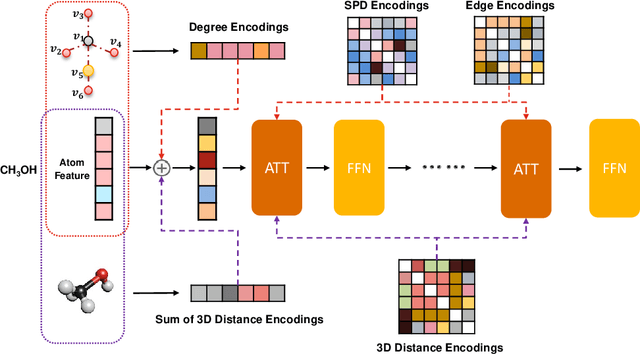
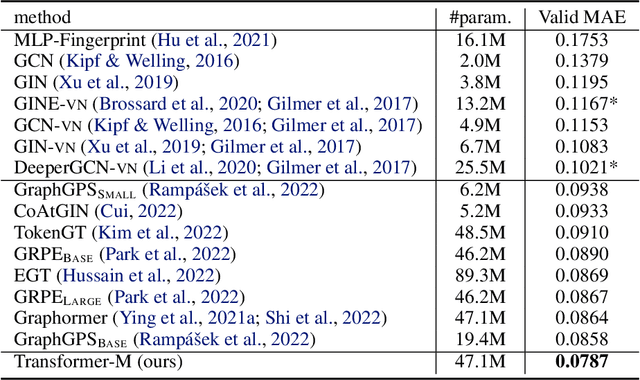
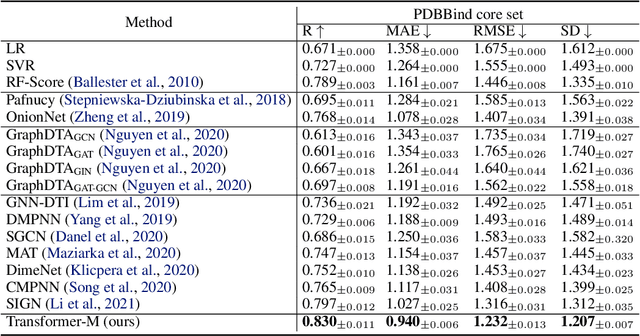
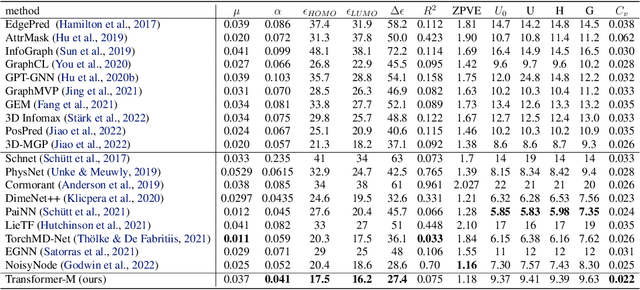
Unlike vision and language data which usually has a unique format, molecules can naturally be characterized using different chemical formulations. One can view a molecule as a 2D graph or define it as a collection of atoms located in a 3D space. For molecular representation learning, most previous works designed neural networks only for a particular data format, making the learned models likely to fail for other data formats. We believe a general-purpose neural network model for chemistry should be able to handle molecular tasks across data modalities. To achieve this goal, in this work, we develop a novel Transformer-based Molecular model called Transformer-M, which can take molecular data of 2D or 3D formats as input and generate meaningful semantic representations. Using the standard Transformer as the backbone architecture, Transformer-M develops two separated channels to encode 2D and 3D structural information and incorporate them with the atom features in the network modules. When the input data is in a particular format, the corresponding channel will be activated, and the other will be disabled. By training on 2D and 3D molecular data with properly designed supervised signals, Transformer-M automatically learns to leverage knowledge from different data modalities and correctly capture the representations. We conducted extensive experiments for Transformer-M. All empirical results show that Transformer-M can simultaneously achieve strong performance on 2D and 3D tasks, suggesting its broad applicability. The code and models will be made publicly available at https://github.com/lsj2408/Transformer-M.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge