Leveraging Dependency Forest for Neural Medical Relation Extraction
Paper and Code
Dec 16, 2019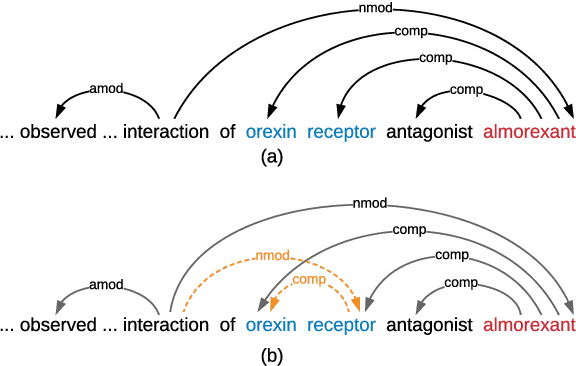
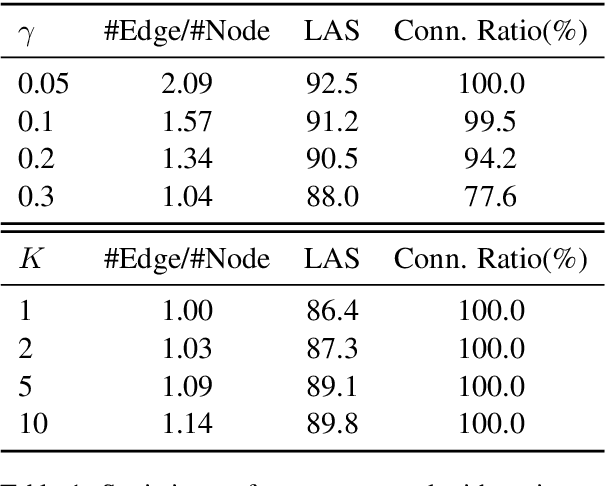
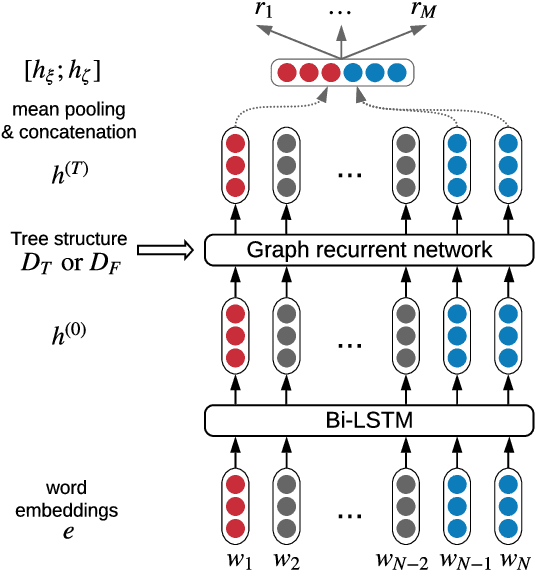
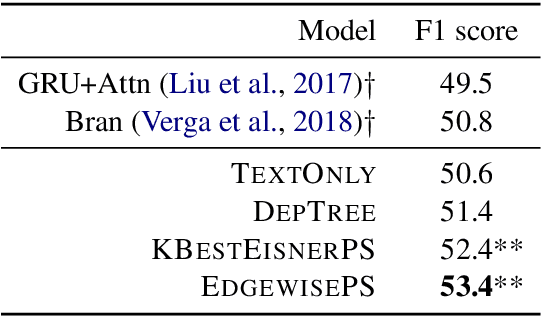
Medical relation extraction discovers relations between entity mentions in text, such as research articles. For this task, dependency syntax has been recognized as a crucial source of features. Yet in the medical domain, 1-best parse trees suffer from relatively low accuracies, diminishing their usefulness. We investigate a method to alleviate this problem by utilizing dependency forests. Forests contain many possible decisions and therefore have higher recall but more noise compared with 1-best outputs. A graph neural network is used to represent the forests, automatically distinguishing the useful syntactic information from parsing noise. Results on two biomedical benchmarks show that our method outperforms the standard tree-based methods, giving the state-of-the-art results in the literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge