Learning to be EXACT, Cell Detection for Asthma on Partially Annotated Whole Slide Images
Paper and Code
Jan 13, 2021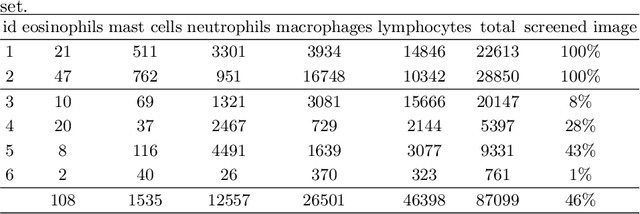
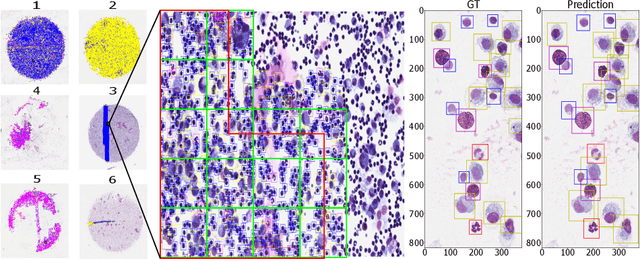
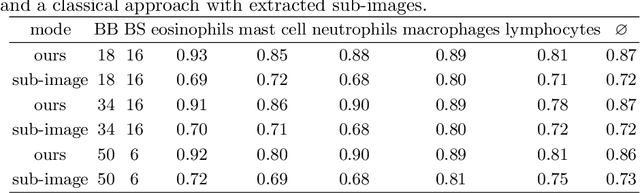
Asthma is a chronic inflammatory disorder of the lower respiratory tract and naturally occurs in humans and animals including horses. The annotation of an asthma microscopy whole slide image (WSI) is an extremely labour-intensive task due to the hundreds of thousands of cells per WSI. To overcome the limitation of annotating WSI incompletely, we developed a training pipeline which can train a deep learning-based object detection model with partially annotated WSIs and compensate class imbalances on the fly. With this approach we can freely sample from annotated WSIs areas and are not restricted to fully annotated extracted sub-images of the WSI as with classical approaches. We evaluated our pipeline in a cross-validation setup with a fixed training set using a dataset of six equine WSIs of which four are partially annotated and used for training, and two fully annotated WSI are used for validation and testing. Our WSI-based training approach outperformed classical sub-image-based training methods by up to 15\% $mAP$ and yielded human-like performance when compared to the annotations of ten trained pathologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge