Latent Diffusion Models for Controllable RNA Sequence Generation
Paper and Code
Sep 15, 2024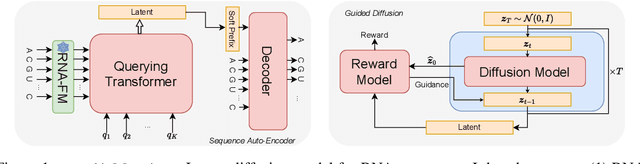
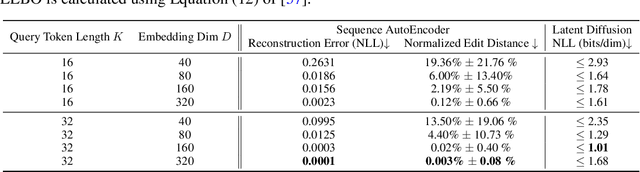

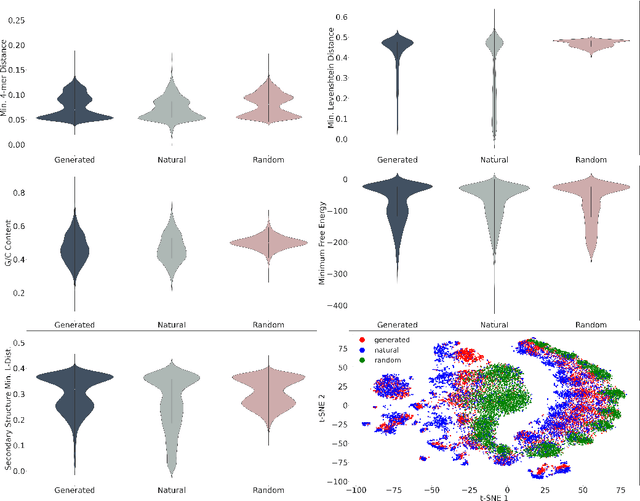
This paper presents RNAdiffusion, a latent diffusion model for generating and optimizing discrete RNA sequences. RNA is a particularly dynamic and versatile molecule in biological processes. RNA sequences exhibit high variability and diversity, characterized by their variable lengths, flexible three-dimensional structures, and diverse functions. We utilize pretrained BERT-type models to encode raw RNAs into token-level biologically meaningful representations. A Q-Former is employed to compress these representations into a fixed-length set of latent vectors, with an autoregressive decoder trained to reconstruct RNA sequences from these latent variables. We then develop a continuous diffusion model within this latent space. To enable optimization, we train reward networks to estimate functional properties of RNA from the latent variables. We employ gradient-based guidance during the backward diffusion process, aiming to generate RNA sequences that are optimized for higher rewards. Empirical experiments confirm that RNAdiffusion generates non-coding RNAs that align with natural distributions across various biological indicators. We fine-tuned the diffusion model on untranslated regions (UTRs) of mRNA and optimize sample sequences for protein translation efficiencies. Our guided diffusion model effectively generates diverse UTR sequences with high Mean Ribosome Loading (MRL) and Translation Efficiency (TE), surpassing baselines. These results hold promise for studies on RNA sequence-function relationships, protein synthesis, and enhancing therapeutic RNA design.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge