InstructBio: A Large-scale Semi-supervised Learning Paradigm for Biochemical Problems
Paper and Code
Apr 14, 2023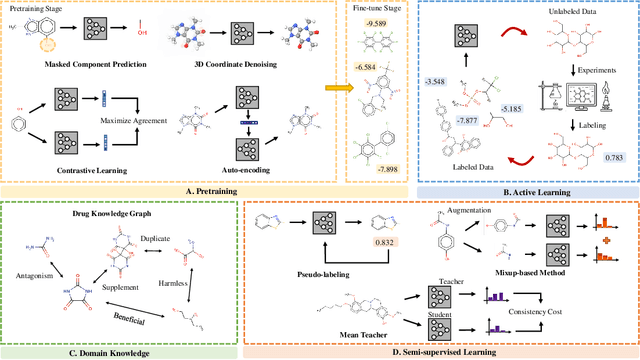
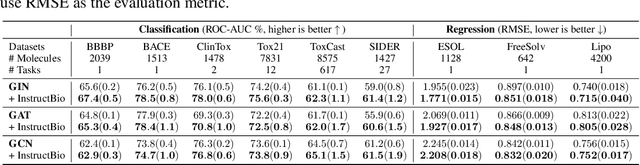
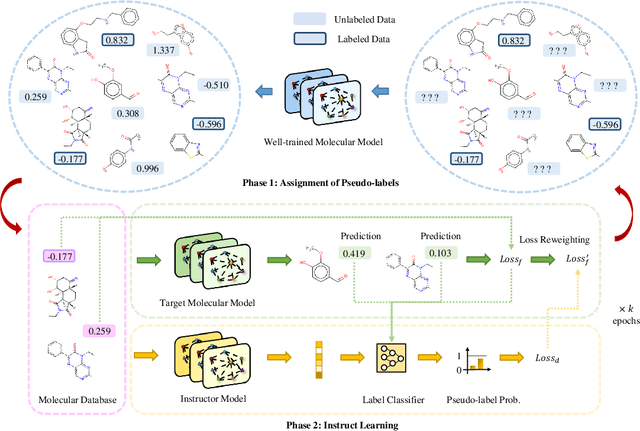
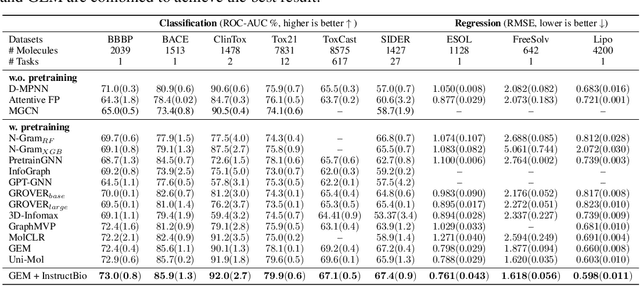
In the field of artificial intelligence for science, it is consistently an essential challenge to face a limited amount of labeled data for real-world problems. The prevailing approach is to pretrain a powerful task-agnostic model on a large unlabeled corpus but may struggle to transfer knowledge to downstream tasks. In this study, we propose InstructMol, a semi-supervised learning algorithm, to take better advantage of unlabeled examples. It introduces an instructor model to provide the confidence ratios as the measurement of pseudo-labels' reliability. These confidence scores then guide the target model to pay distinct attention to different data points, avoiding the over-reliance on labeled data and the negative influence of incorrect pseudo-annotations. Comprehensive experiments show that InstructBio substantially improves the generalization ability of molecular models, in not only molecular property predictions but also activity cliff estimations, demonstrating the superiority of the proposed method. Furthermore, our evidence indicates that InstructBio can be equipped with cutting-edge pretraining methods and used to establish large-scale and task-specific pseudo-labeled molecular datasets, which reduces the predictive errors and shortens the training process. Our work provides strong evidence that semi-supervised learning can be a promising tool to overcome the data scarcity limitation and advance molecular representation learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge