Improving Robustness of Deep Learning Based Knee MRI Segmentation: Mixup and Adversarial Domain Adaptation
Paper and Code
Aug 14, 2019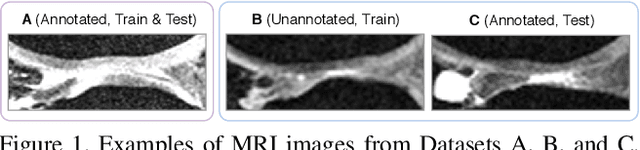
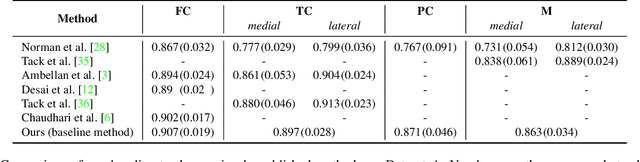
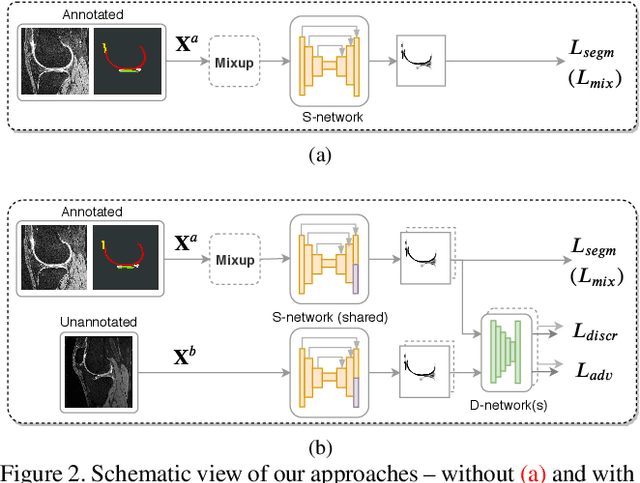
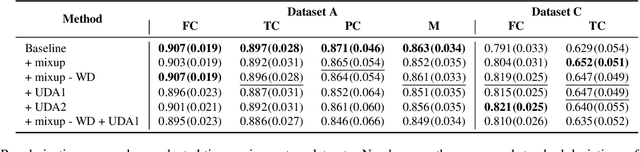
Degeneration of articular cartilage (AC) is actively studied in knee osteoarthritis (OA) research via magnetic resonance imaging (MRI). Segmentation of AC tissues from MRI data is an essential step in quantification of their damage. Deep learning (DL) based methods have shown potential in this realm and are the current state-of-the-art, however, their robustness to heterogeneity of MRI acquisition settings remains an open problem. In this study, we investigated two modern regularization techniques -- mixup and adversarial unsupervised domain adaptation (UDA) -- to improve the robustness of DL-based knee cartilage segmentation to new MRI acquisition settings. Our validation setup included two datasets produced by different MRI scanners and using distinct data acquisition protocols. We assessed the robustness of automatic segmentation by comparing mixup and UDA approaches to a strong baseline method at different OA severity stages and, additionally, in relation to anatomical locations. Our results showed that for moderate changes in knee MRI data acquisition settings both approaches may provide notable improvements in the robustness, which are consistent for all stages of the disease and affect the clinically important areas of the knee joint. However, mixup may be considered as a recommended approach, since it is more computationally efficient and does not require additional data from the target acquisition setup.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge