Hyperspectral band selection using genetic algorithm and support vector machines for early identification of charcoal rot disease in soybean
Paper and Code
Oct 12, 2017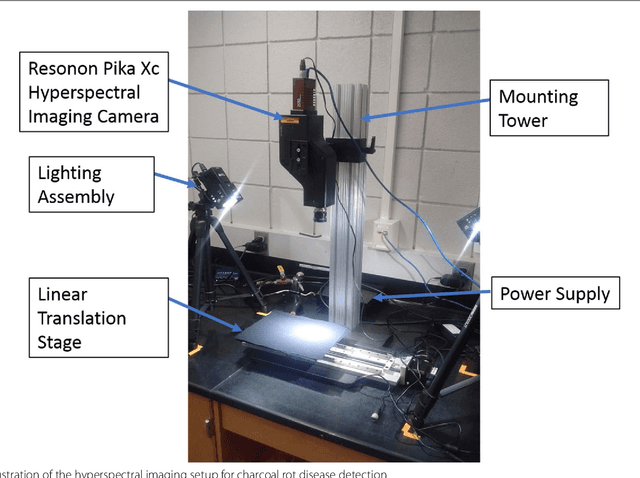



Charcoal rot is a fungal disease that thrives in warm dry conditions and affects the yield of soybeans and other important agronomic crops worldwide. There is a need for robust, automatic and consistent early detection and quantification of disease symptoms which are important in breeding programs for the development of improved cultivars and in crop production for the implementation of disease control measures for yield protection. Current methods of plant disease phenotyping are predominantly visual and hence are slow and prone to human error and variation. There has been increasing interest in hyperspectral imaging applications for early detection of disease symptoms. However, the high dimensionality of hyperspectral data makes it very important to have an efficient analysis pipeline in place for the identification of disease so that effective crop management decisions can be made. The focus of this work is to determine the minimal number of most effective hyperspectral bands that can distinguish between healthy and diseased specimens early in the growing season. Healthy and diseased hyperspectral data cubes were captured at 3, 6, 9, 12, and 15 days after inoculation. We utilized inoculated and control specimens from 4 different genotypes. Each hyperspectral image was captured at 240 different wavelengths in the range of 383 to 1032 nm. We used a combination of genetic algorithm as an optimizer and support vector machines as a classifier for identification of maximally effective band combinations. A binary classification between healthy and infected samples using six selected band combinations obtained a classification accuracy of 97% and a F1 score of 0.97 for the infected class. The results demonstrated that these carefully chosen bands are more informative than RGB images, and could be used in a multispectral camera for remote identification of charcoal rot infection in soybean.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge