GaKCo: a Fast GApped k-mer string Kernel using COunting
Paper and Code
Sep 18, 2017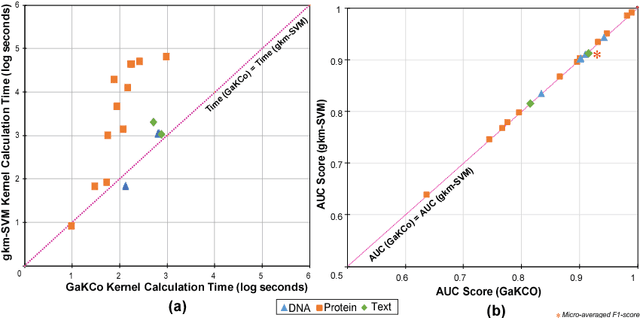
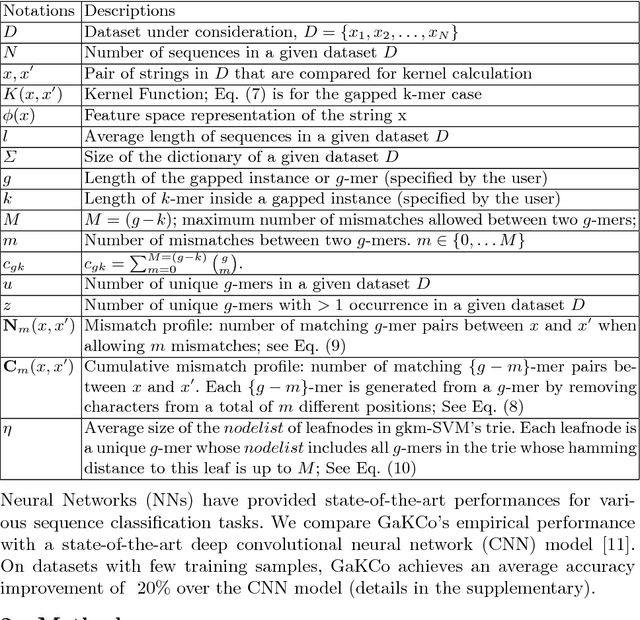
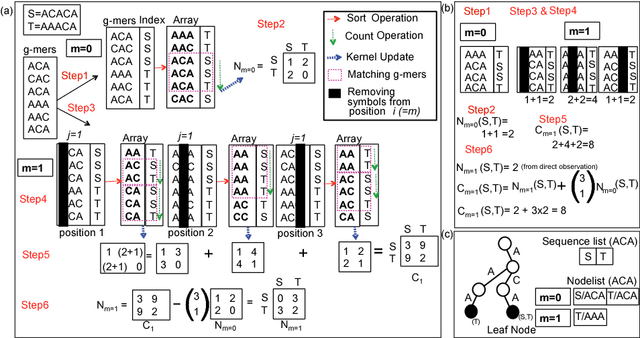
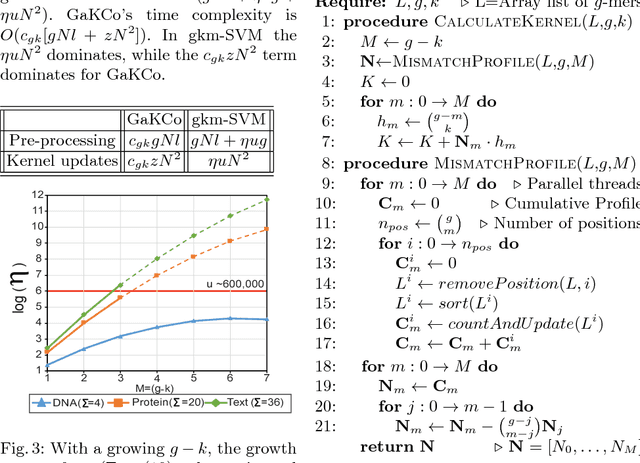
String Kernel (SK) techniques, especially those using gapped $k$-mers as features (gk), have obtained great success in classifying sequences like DNA, protein, and text. However, the state-of-the-art gk-SK runs extremely slow when we increase the dictionary size ($\Sigma$) or allow more mismatches ($M$). This is because current gk-SK uses a trie-based algorithm to calculate co-occurrence of mismatched substrings resulting in a time cost proportional to $O(\Sigma^{M})$. We propose a \textbf{fast} algorithm for calculating \underline{Ga}pped $k$-mer \underline{K}ernel using \underline{Co}unting (GaKCo). GaKCo uses associative arrays to calculate the co-occurrence of substrings using cumulative counting. This algorithm is fast, scalable to larger $\Sigma$ and $M$, and naturally parallelizable. We provide a rigorous asymptotic analysis that compares GaKCo with the state-of-the-art gk-SK. Theoretically, the time cost of GaKCo is independent of the $\Sigma^{M}$ term that slows down the trie-based approach. Experimentally, we observe that GaKCo achieves the same accuracy as the state-of-the-art and outperforms its speed by factors of 2, 100, and 4, on classifying sequences of DNA (5 datasets), protein (12 datasets), and character-based English text (2 datasets), respectively. GaKCo is shared as an open source tool at \url{https://github.com/QData/GaKCo-SVM}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge